

植物学报 ›› 2016, Vol. 51 ›› Issue (2): 202-209.DOI: 10.11983/CBB15088 cstr: 32102.14.CBB15088
李冬梅1, 王路雅1, 张澜玥2, 帖子阳2, 毛惠平1,*( )
)
收稿日期:2015-05-18
接受日期:2015-09-04
出版日期:2016-03-01
发布日期:2016-03-31
通讯作者:
E-mail: 基金资助:
Dongmei Li1, Luya Wang1, Lanyue Zhang2, Ziyang Tie2, Huiping Mao1,*( )
)
Received:2015-05-18
Accepted:2015-09-04
Online:2016-03-01
Published:2016-03-31
Contact:
E-mail: 摘要: AtPROPEP是拟南芥(Arabidopsis thaliana)具有7个成员的基因家族, 编码内源短肽激素。AtPROPEP基因家族编码的蛋白质C端23个氨基酸短肽能够被2个同源激酶受体AtPEPR1和AtPEPR2识别并结合, 引起下游反应。然而, 对于该家族成员AtPROPEP2,3−6的表达对茉莉酸(JA)和水杨酸(SA)的响应以及在根生长中的作用并不清楚。GUS染色和定量RT-PCR分析结果表明, AtPROPEP2-6的表达对于JA和SA的响应不同, 暗示着它们可能通过不同的方式参与植物的先天免疫反应。AtPROPEP3和AtPROPEP4过表达植株的表型分析表明, AtPROPEP3和AtPROPEP4促进拟南芥根的生长。
李冬梅, 王路雅, 张澜玥, 帖子阳, 毛惠平. 拟南芥短肽激素PROPEP基因家族在根生长中的作用机理. 植物学报, 2016, 51(2): 202-209.
Dongmei Li, Luya Wang, Lanyue Zhang, Ziyang Tie, Huiping Mao. Mechanism of Arabidopsis Short Peptide Hormones PROPEP Gene Family in the Root Growth. Chinese Bulletin of Botany, 2016, 51(2): 202-209.
| Primer name | Sequences (5'-3') | Function |
|---|---|---|
| AtPROPEP1_PF-HindIII AtPROPEP1_PR-BamHI AtPROPEP2_PF-HindIII AtPROPEP2_PR-BamHI AtPROPEP3_PF-BamHI AtPROPEP3_PR-NotI AtPROPEP4_PF-HindIII AtPROPEP4_PR-EcoRI AtPROPEP5_PF-HindIII AtPROPEP5_PR-BamHI AtPROPEP6_PF-XhoI AtPROPEP6_PR-EcoRI | CCCAAGCTTGTAAATTATAGTGAAAGGTACGG GCGGATCCTGAGATCTGATAAGACAGAGG CCCAAGCTTCGCATTCGCTTTTTTCTTTTTG GCGGATCCTGAAATCCAATAGTTTGTGAG GCGGATCCTATTTTAACAGTCAACAGCTATTTGG TTGCGGCCGCCGTTGACTTCTTAATCTTTTTTTG CCCAAGCTTAATAAGGATGAATAAAAAGTTTGGG CCGGAATTCGTTTTTCTTCAATTCTGCTTCGTG CCCAAGCTTTACTTAATTTCTTGTGAGAAACTTG GCGGATCCCTTCGCTATCTTCTAAGTTCCTC CCCTCGAGTGATATCTAAGTCCAACTTGGTG CCGGAATTCGTTTTTTGTTTTCTTTCTCTTCTT | AtPROPEP1 promoter clone AtPROPEP2 promoter clone AtPROPEP3 promoter clone AtPROPEP4 promoter clone AtPROPEP5 promoter clone AtPROPEP6 promoter clone |
表1 AtPROPEPs启动子克隆引物序列
Table 1 Sequences of AtPROPEPs promoter clone primers
| Primer name | Sequences (5'-3') | Function |
|---|---|---|
| AtPROPEP1_PF-HindIII AtPROPEP1_PR-BamHI AtPROPEP2_PF-HindIII AtPROPEP2_PR-BamHI AtPROPEP3_PF-BamHI AtPROPEP3_PR-NotI AtPROPEP4_PF-HindIII AtPROPEP4_PR-EcoRI AtPROPEP5_PF-HindIII AtPROPEP5_PR-BamHI AtPROPEP6_PF-XhoI AtPROPEP6_PR-EcoRI | CCCAAGCTTGTAAATTATAGTGAAAGGTACGG GCGGATCCTGAGATCTGATAAGACAGAGG CCCAAGCTTCGCATTCGCTTTTTTCTTTTTG GCGGATCCTGAAATCCAATAGTTTGTGAG GCGGATCCTATTTTAACAGTCAACAGCTATTTGG TTGCGGCCGCCGTTGACTTCTTAATCTTTTTTTG CCCAAGCTTAATAAGGATGAATAAAAAGTTTGGG CCGGAATTCGTTTTTCTTCAATTCTGCTTCGTG CCCAAGCTTTACTTAATTTCTTGTGAGAAACTTG GCGGATCCCTTCGCTATCTTCTAAGTTCCTC CCCTCGAGTGATATCTAAGTCCAACTTGGTG CCGGAATTCGTTTTTTGTTTTCTTTCTCTTCTT | AtPROPEP1 promoter clone AtPROPEP2 promoter clone AtPROPEP3 promoter clone AtPROPEP4 promoter clone AtPROPEP5 promoter clone AtPROPEP6 promoter clone |
| Primer name | Sequences (5'-3') | Function |
|---|---|---|
| AtPROPEP3_CDSF-SalI AtPROPEP3_CDSR-BamHI AtPROPEP4_CDSF-SalI AtPROPEP4_CDSR-EcoRI | GCGTCGACATGGAGAATCTCAGAAATGG CGGGATCCCTAATTGTGTTTGCCTCCTT GCGTCGACATGGAGAGAGGAGTTTCTTA CGGAATTCCTAAAACGGCTTCTTGTTGG | AtPROPEP3 coding sequence clone AtPROPEP4 coding sequence clone |
表2 AtPROPEP3和AtPROPEP4引物序列
Table 2 Sequences of AtPROPEP3 and AtPROPEP4 clone primers
| Primer name | Sequences (5'-3') | Function |
|---|---|---|
| AtPROPEP3_CDSF-SalI AtPROPEP3_CDSR-BamHI AtPROPEP4_CDSF-SalI AtPROPEP4_CDSR-EcoRI | GCGTCGACATGGAGAATCTCAGAAATGG CGGGATCCCTAATTGTGTTTGCCTCCTT GCGTCGACATGGAGAGAGGAGTTTCTTA CGGAATTCCTAAAACGGCTTCTTGTTGG | AtPROPEP3 coding sequence clone AtPROPEP4 coding sequence clone |
| Primer name | Sequences (5'-3') |
|---|---|
| AtPROPEP2F AtPROPEP2R AtPROPEP3F AtPROPEP3R AtPROPEP4F AtPROPEP4R AtPROPEP5F AtPROPEP5R AtPROPEP6F AtPROPEP6R ACTINF ACTINR | CTCGACCAAGCTCTCATAGCTG CACAACGACATCATCGTCTTTC TCTTCTTCTTGCGATCTTTCGTCAT CTGAACTTGGCGTAGGCTTAGTC CTCAAGCTTCTCGGTTTGCGATC ACTTTCTCTCGACTTCTTTAGTAC GAGATTGTTGCAAGCTCATGCCTC AGTTGAAGTTTCGATAGATGAAGGT TGAAGTGTCTTGGTCTTGAGTC TGGTCTCCTTCTTAACACTGCTG ACGGTAACATTGTGCTCAGTGGTG CTTGGAGATCCACATCTGCTGGA |
表3 实时荧光定量PCR引物
Table 3 Primers used for quantitative RT-PCR
| Primer name | Sequences (5'-3') |
|---|---|
| AtPROPEP2F AtPROPEP2R AtPROPEP3F AtPROPEP3R AtPROPEP4F AtPROPEP4R AtPROPEP5F AtPROPEP5R AtPROPEP6F AtPROPEP6R ACTINF ACTINR | CTCGACCAAGCTCTCATAGCTG CACAACGACATCATCGTCTTTC TCTTCTTCTTGCGATCTTTCGTCAT CTGAACTTGGCGTAGGCTTAGTC CTCAAGCTTCTCGGTTTGCGATC ACTTTCTCTCGACTTCTTTAGTAC GAGATTGTTGCAAGCTCATGCCTC AGTTGAAGTTTCGATAGATGAAGGT TGAAGTGTCTTGGTCTTGAGTC TGGTCTCCTTCTTAACACTGCTG ACGGTAACATTGTGCTCAGTGGTG CTTGGAGATCCACATCTGCTGGA |
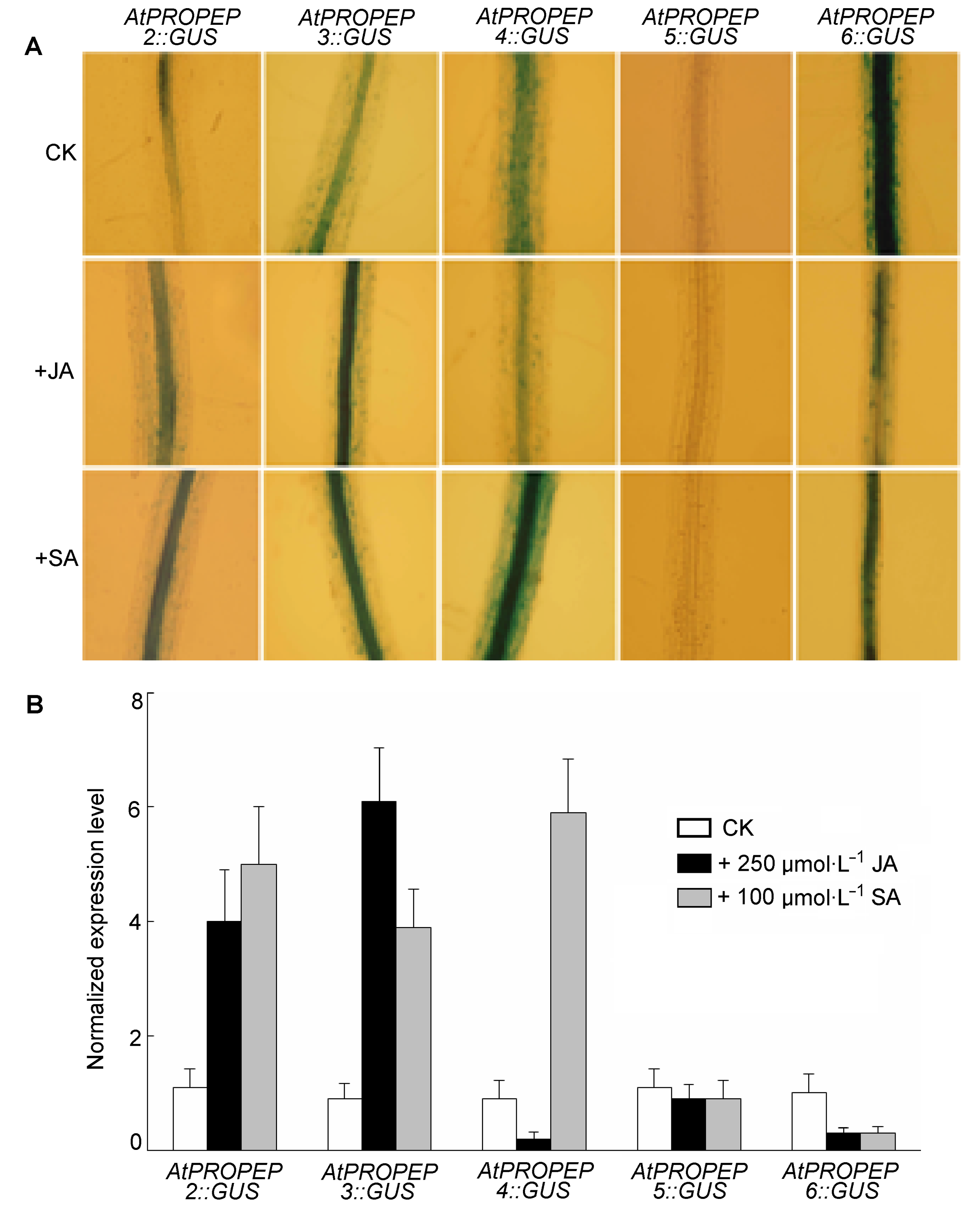
图2 AtPROPEP2,3−6启动子对茉莉酸(JA)和水杨酸(SA)的响应 (A) AtPROPEP2,3−6::GUS转基因植株在250 µmol∙L−1 JA和100 µmol∙L−1 SA处理下的GUS染色; (B) 定量RT-PCR检测AtPROPEP2,3−6在JA和SA处理下的表达水平
Figure 2 Response of AtPROPEP2,3−6 promoter to jasmonic acid (JA) and salicylic acid (SA) (A) GUS staining of transgenic seedlings harboring AtPROPEP2,3−6::GUS construct under 250 µmol∙L−1 JA and 100 µmol∙L−1 SA; (B) Quantitative RT-PCR analysis of AtPROPEP2,3−6 expression level under JA and SA treatments
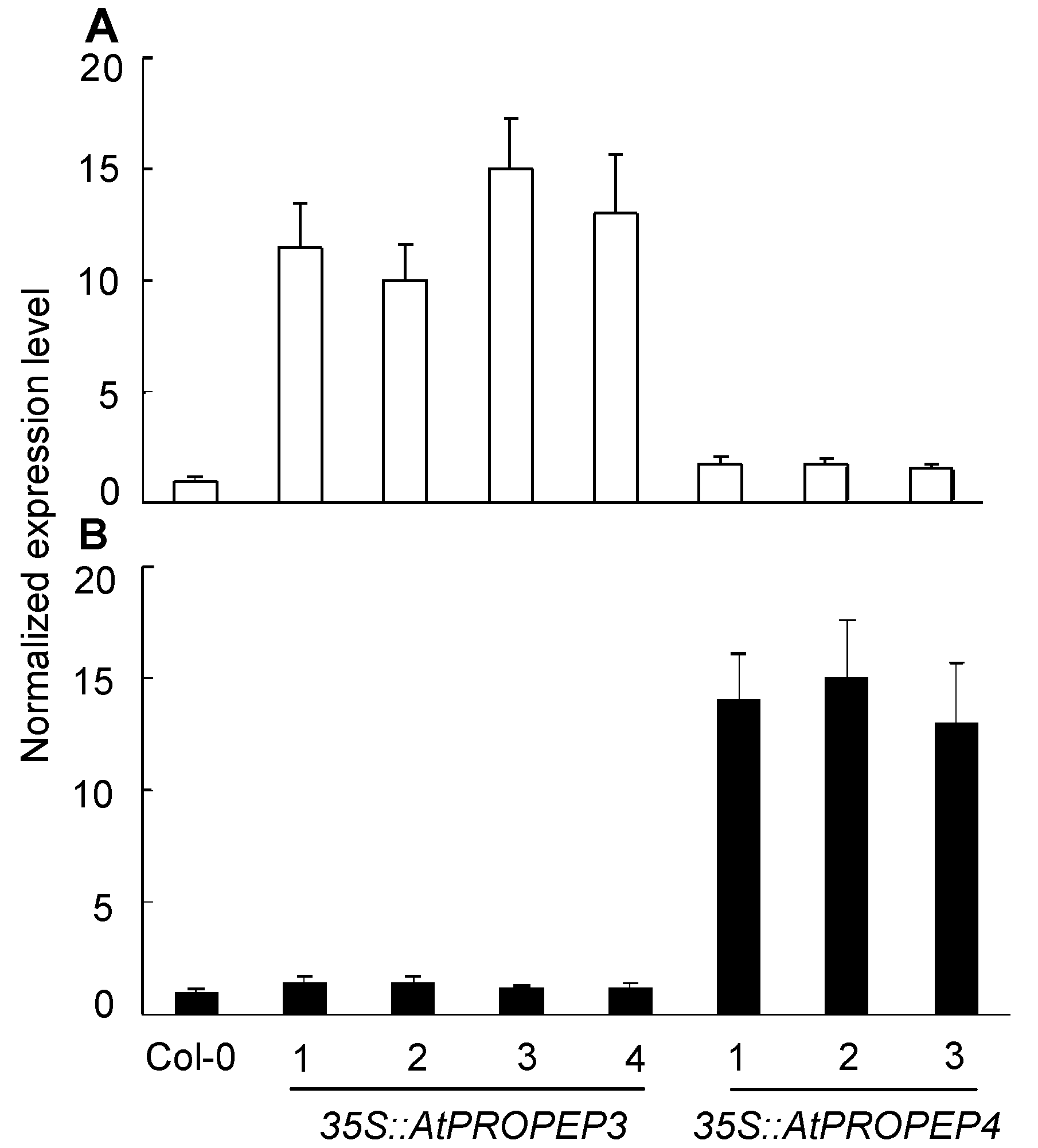
图3 定量RT-PCR检测拟南芥过表达植株 (A) 35S::AtPROPEP3; (B) 35S::AtPROPEP4
Figure 3 Quantitative RT-PCR analysis of Arabidopsis over- expressing lines (A) 35S::AtPROPEP3; (B) 35S::AtPROPEP4
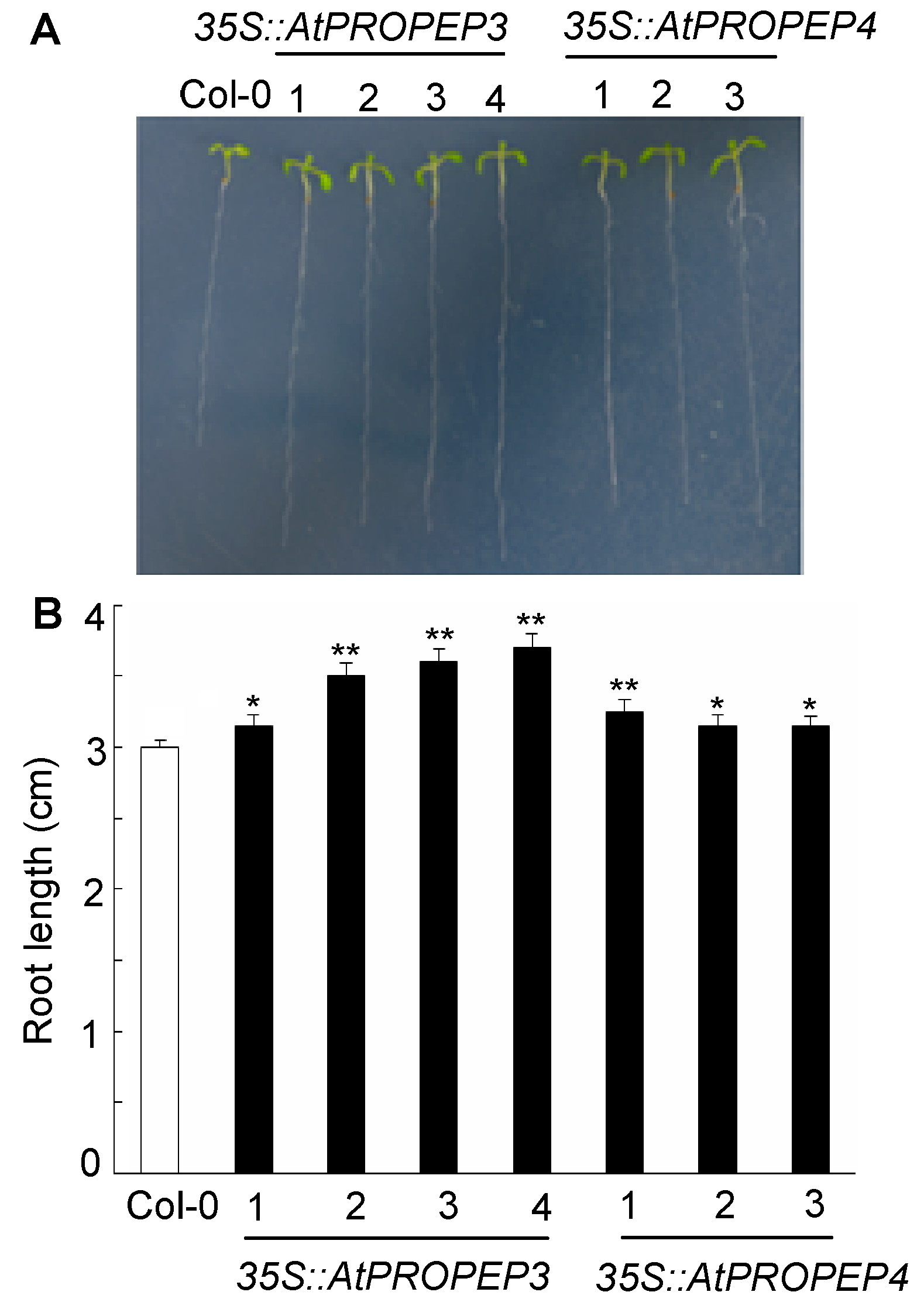
图4 拟南芥过量表达AtPROPEP3和AtPROPEP4植株根长增加 (A) 野生型与过量表达突变体在CK培养基上的表型; (B) 野生型与过量表达突变体在CK培养基上的根长分析(**P<0.01; * P<0.05)
Figure 4 Arabidopsis plants overexpressing AtPROPEP3 and AtPROPEP4 exhibit longer root than that of wild type (A) Seedlings of Col-0 and transgenic seedlings grown on CK medium; (B) Measurement of root growth under CK medium (**P<0.01; * P<0.05)
| [1] | 李新锋, 赵淑清 (2004). 转基因植物中报告基因GUS的活性检测及其应用. 生命的化学 24, 71-74. |
| [2] | Bartels S, Lori M, Mbengue M, van Verk M, Klauser D, Hander T, Böni R, Robatzek S, Boller T (2013). The family of Peps and their precursors in Arabidopsis: dif- ferential expression and localization but similar induction of pattern-triggered immune responses. J Exp Bot 64, 5309-5321. |
| [3] | Gijrlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, Ryals J (1996). Bekothiadiazdle, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8, 629-643. |
| [4] |
Huffaker A, Pearce G, Ryan CA (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103, 10098-10103.
DOI PMID |
| [5] |
Huffaker A, Ryan CA (2007). Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA 104, 10732-10736.
DOI PMID |
| [6] | Kuc J (1982). Induced immunity to plant disease. Bioscience 32, 854-860. |
| [7] | Ma C, Guo J, Kang Y, Doman K, Bryan AC, Tax FE, Yamaguchi Y, Qi Z (2014). AtPEPTIDE RECEPTOR2 mediates the AtPEPTIDE1 induced cytosolic Ca2+ rise which is required for the suppression of Glutamate Dumper gene expression in Arabidopsis roots. J Integr Plant Biol 56, 684-694. |
| [8] | Pearce G, Moura DS, Stratmann J, Ryan CA (2001). Production of multiple plant hormones from a single polyprotein precursor. Nature 411, 817-820. |
| [9] |
Ross AF (1961). Systemic acquired resistance induced by localized virus infections in plants. Virology 14, 340-358.
PMID |
| [10] |
Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645-656.
DOI PMID |
| [11] | Van Loon LC, Van Kammen A (1970). Polyacrylamide disc electrophoresis of the soluble proteins from Nicotiene tabacum var. samsun and Samnkun NN. II. Changes in protein constitution after infection with tobacco mosaic virus. Virology 40, 199-211. |
| [12] |
Vijayan P, Shockey J, Lévesque CA, Cook RJ, Browse J (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc Natl Acad Sci USA 95, 7209-7214.
DOI PMID |
| [13] | Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA (1991). Co-ordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085-1094. |
| [14] | Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508-522. |
| [15] | Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip methods. Nature Protocols 1, 641-646. |
| [16] |
Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Some- rville S (2004). Host and non-host pathogens elicit differ- ent jasmonate/ethylene responses in Arabidopsis. Plant J 40, 633-646.
DOI PMID |
| [1] | 张子睿, 周静, 胡艳萍, 梁爽, 马永鹏, 陈伟乐. 极度濒危植物巧家五针松的根内和根际真菌群落特征[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | 王秀媛, 申磊, 刘婷婷, 尉雯雯, 张帅, 张伟. ‘塞外红’苹果-大豆复合系统根系时空分布与种间竞争策略[J]. 植物生态学报, 2025, 49(5): 748-759. |
| [3] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| [4] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [5] | 王娟, 张登山, 肖元明, 裴全帮, 王博, 樊博, 周国英. 长期围封后高寒草原植物根系分泌物特征与环境因子关系[J]. 植物生态学报, 2025, 49(4): 596-609. |
| [6] | 宋威, 程才, 王嘉伟, 吴纪华. 土壤微生物对植物多样性-生态系统功能关系的调控作用[J]. 生物多样性, 2025, 33(4): 24579-. |
| [7] | 李梦琦, 苗灵凤, 李大东, 龙奕帆, 叶冰冰, 杨帆. 海南东寨港红树林植物细根功能性状对不同潮位沉积物养分变化的响应[J]. 植物生态学报, 2025, 49(4): 552-561. |
| [8] | 刘雨函, 曹启江, 张诗晗, 李益慧, 王菁, 谭晓萌, 刘筱儒, 王显玲. 拟南芥AtFTCD-L参与根系响应土壤紧实度的机制研究[J]. 植物学报, 2025, 60(4): 1-0. |
| [9] | 杜英杰, 范爱连, 王雪, 闫晓俊, 陈廷廷, 贾林巧, 姜琦, 陈光水. 亚热带天然常绿阔叶林乔木树种与林下灌木树种根-叶功能性状协调性及差异[J]. 植物生态学报, 2025, 49(4): 585-595. |
| [10] | 田伟, 陈佳杰, 陈渊戈, 徐清, 周进. 浙江象山港蟹类(十足目: 短尾下目)物种多样性[J]. 生物多样性, 2025, 33(4): 24461-. |
| [11] | 郭李琦, 闫晓蕾, 曹磊, 高景, 刘瑞强, 周旭辉. 树种菌根类型与根系性状对根际微生物网络复杂性的影响[J]. 植物生态学报, 2025, 49(4): 573-584. |
| [12] | 曾文丹, 严华兵, 吴正丹, 尚小红, 曹升, 陆柳英, 肖亮, 施平丽, 程冬, 龙紫媛, 李婕宇. 发根农杆菌介导的野葛毛状根遗传转化体系[J]. 植物学报, 2025, 60(3): 425-434. |
| [13] | 杨莉, 曲茜彤, 陈子航, 邹婷婷, 王全华, 王小丽. 菠菜AT-hook基因家族鉴定与表达谱分析[J]. 植物学报, 2025, 60(3): 377-392. |
| [14] | 郑琳敏, 熊小玲, 姜永孟, 王曼, 张锦秀, 曾志伟, 吕茂奎, 谢锦升. 武夷山不同海拔杉木凋落叶和细根分解规律以及驱动因素的差异[J]. 植物生态学报, 2025, 49(2): 244-255. |
| [15] | 李建国, 张怡, 张文君. 水稻根系铁膜形成及对磷吸收的影响[J]. 植物学报, 2025, 60(1): 132-143. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||