

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (5): 625-633.DOI: 10.11983/CBB17169
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Liu Wei1, Tong Yong’ao1,2, Bai Jie1,*( )
)
Received:2017-09-06
Accepted:2017-11-16
Online:2018-09-01
Published:2018-11-29
Contact:
Bai Jie
About author:† These authors contributed equally to this paper
Liu Wei, Tong Yong’ao, Bai Jie. Bioinformatics Analysis of tRNA-derived Fragments in Rice Male Gametophyte Development[J]. Chinese Bulletin of Botany, 2018, 53(5): 625-633.
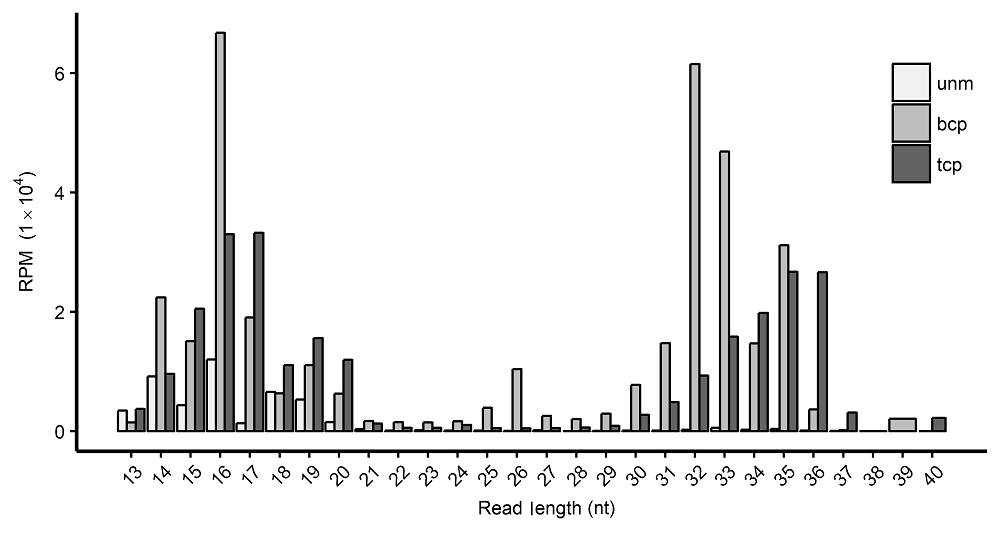
Figure 1 Length distribution of tRFs in three types of pollens in rice male gameteunm: Uni-nucleate microspore; bcp: Bi-cellular pollen; tcp : Tri-cellular pollen; RPM: Reads per million
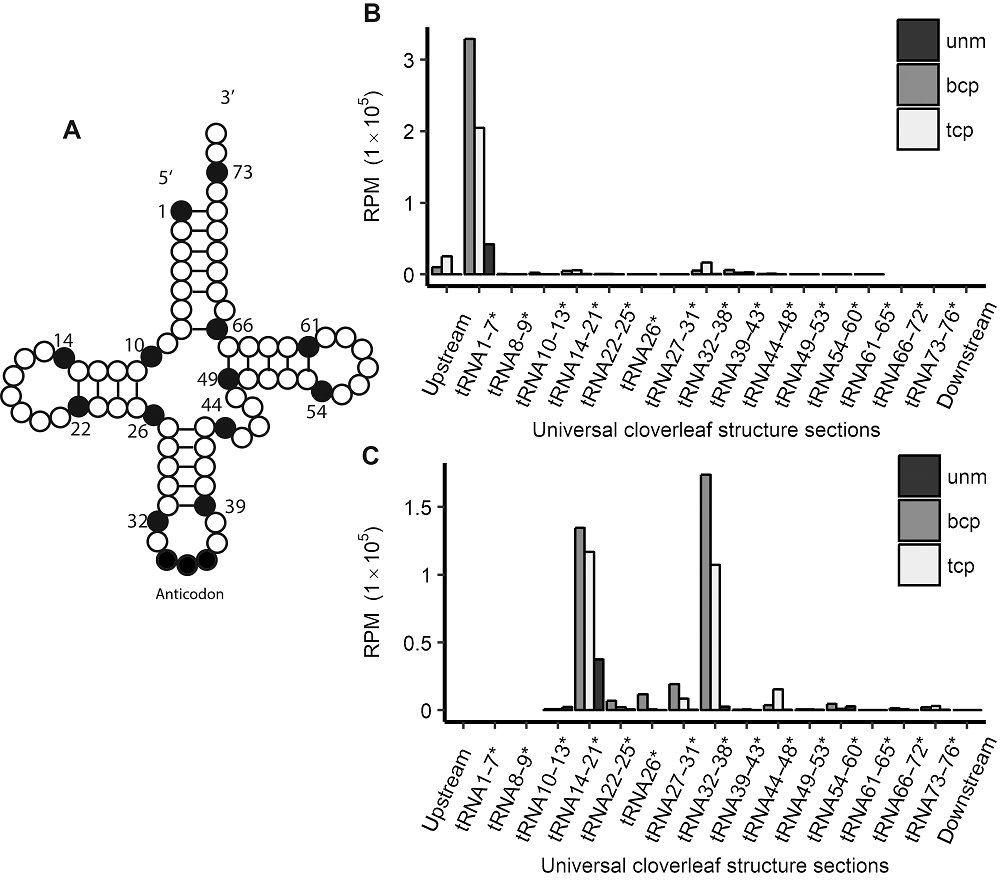
Figure 2 tRFs’ starting and ending site distribution in tRNAs’ universal cloverleaf structure sections in three types of pollens in rice male gamete(A) Schematic view of tRNAs’ cloverleaf-like secondary structure; (B) tRFs’ starting site positions in tRNA; (C) tRFs’ ending site positions in tRNA. * indicate that the numbers before can only be regarded as a positions represented in Figure 2A instead of base ID. unm, bcp, tcp and RPM see Figure 1.
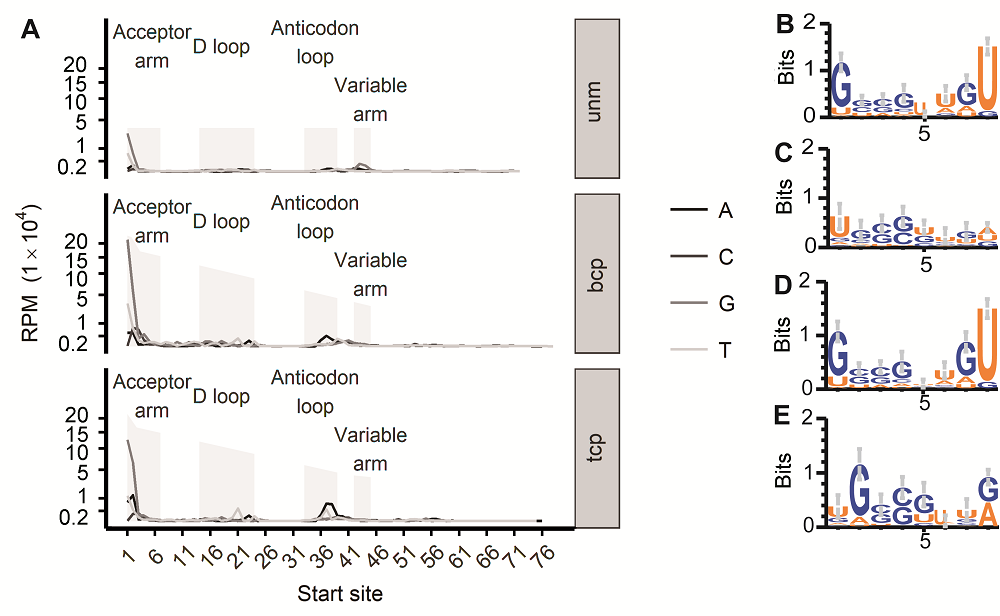
Figure 3 tRFs’ 5’ end base bias in three types of pollens in rice male gamete(A) tRF starting site’s distribution in each position of their host tRNA (The vertical axis labels’ positions were transferred according to their square root number); (B) First 8 base logo of the tRNAs that produce tRFs from the 1st base in bcp; (C) First 8 base logo of tRNAs that produce tRFs from the 2nd base in bcp; (D) First 8 base logo of the tRNAs that produce tRFs from the 1st base in tcp; (E) First 8 base logo of tRNAs that produce tRFs from the 2nd base in tcp. unm, bcp, tcp and RPM see Figure 1.
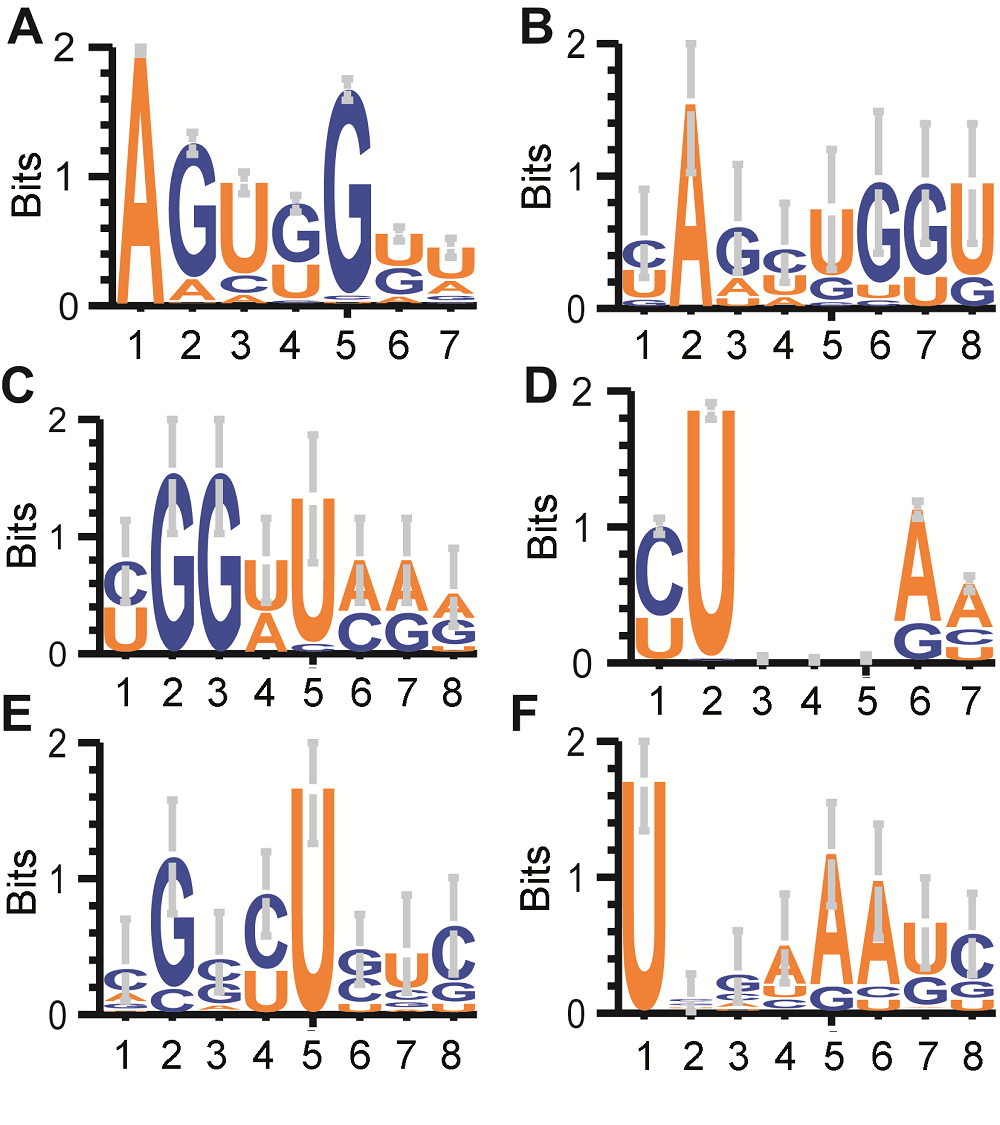
Figure 4 Sequence motif logo for tRNA 8 mer around cleavage sites in three types of pollens in rice male gamete(A), (D) Sequence logo of all tRNA’s first 7 bases of D loop and Anticodon loop; (B) For bcp tRFs ended at the 3rd base of D loop; (C) For tcp tRFs started at the 7th base of D loop; (E) For bcp tRFs ended at the 1st base of Anticodon loop; (F) For tcp tRFs started at the 6th base of Anticodon loop. The position between 4 and 5 in X axis of Figure B, C, E and F stand for cleavage site.
| Sample | No. of non-TE genes | No. of genic TEs | No. of intergenic TEs | TE Percentage (%) |
|---|---|---|---|---|
| unm | 317 | 166 | 506 | 67.95 |
| bcp | 728 | 403 | 905 | 64.24 |
| tcp | 529 | 287 | 736 | 65.91 |
Table 1 tRFs targeted loci of non-TE and TE genes
| Sample | No. of non-TE genes | No. of genic TEs | No. of intergenic TEs | TE Percentage (%) |
|---|---|---|---|---|
| unm | 317 | 166 | 506 | 67.95 |
| bcp | 728 | 403 | 905 | 64.24 |
| tcp | 529 | 287 | 736 | 65.91 |
| 1 | Abe T, Ikemura T, Ohara Y, Uehara H, Kinouchi M, Kanaya S, Yamada Y, Muto A, Inokuchi H (2009). tRNADB-CE: tRNA gene database curated manually by experts.Nucleic Acids Res 37, D163-D168. |
| 2 | Alves CS, Vicentini R, Duarte GT, Pinoti VF, Vincentz M, Nogueira FTS (2016). Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants.Plant Mol Biol 93, 35-48. |
| 3 | Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou XP, Russell SD, Sundaresan V (2013). Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J 76, 729-741. |
| 4 | Chen CJ, Liu Q, Zhang YC, Qu LH, Chen YQ, Gautheret D (2011). Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus.RNA Biol 8, 538-547. |
| 5 | Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS, Green PJ, Barton GJ, Hutvagner G (2009). Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs.RNA 15, 2147-2160. |
| 6 | Couvillion MT, Sachidanandam R, Collins K (2010). A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 24, 2742-2747. |
| 7 | Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM (2006). MODOMICS: a database of RNA modification pathways.Nucleic Acids Res 34, D145-D149. |
| 8 | Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P (2010). Angiogenin- induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly.J Biol Chem 285, 10959-10968. |
| 9 | Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N (2017). A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress.RNA Biol 14, 1364-1373. |
| 10 | Goodarzi H, Liu XH, Nguyen HCB, Zhang S, Fish L, Tavazoie SF (2015). Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement.Cell 161, 790-802. |
| 11 | Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H (2009). MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 10, 643. |
| 12 | Hsieh LC, Lin SI, Kuo HF, Chiou TJ (2010). Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots.Plant Signal Behav 5, 537-539. |
| 13 | Karaiskos S, Naqvi AS, Swanson KE, Grigoriev A (2015). Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol Direct 10, 51. |
| 14 | Keam SP, Sobala A, Ten Have S, Hutvagner G (2017). tRNA-Derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation.J Proteome Res 16, 413-420. |
| 15 | Kumar P, Anaya J, Mudunuri SB, Dutta A (2014). Meta- analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets.BMC Biol 12, 78. |
| 16 | Kumar P, Kuscu C, Dutta A (2016). Biogenesis and function of transfer RNA-related fragments (tRFs).Trends Biochem Sci 41, 679-689. |
| 17 | Loss-Morais G, Waterhouse PM, Margis R (2013). Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets.Biol Direct 8, 6. |
| 18 | Lowe TM, Eddy SR (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence.Nucleic Acids Res 25, 955-964. |
| 19 | Martinez G, Choudury SG, Slotkin RK (2017). tRNA- derived small RNAs target transposable element transcripts.Nucleic Acids Res 45, 5142-5152. |
| 20 | Nawrot B, Malkiewicz A, Smith WS, Sierzputowska-Gracz H, Agris PF (1995). RNA modified uridines VII: chemical synthesis and initial analysis of tRNA D-Loop oligomers with tandem modified uridines.Nucleos Nucleot Nud 14, 143-165. |
| 21 | Peng H, Chun J, Ai TB, Tong YA, Zhang R, Zhao MM, Chen F, Wang SH (2012). MicroRNA profiles and their control of male gametophyte development in rice.Plant Mol Biol 80, 85-102. |
| 22 | Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F (2010). RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage.Genes Dev 24, 1590-1595. |
| 23 | Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun FY, Song LN, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals.Science 351, 391-396. |
| 24 | Slotkin RK, Vaughn M, Borges F, Tanurdžić M, Becker JD, Feijo JA, Martienssen RA (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen.Cell 136, 461-472. |
| 25 | Sobala A, Hutvagner G (2013). Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells.RNA Biol 10, 553-563. |
| 26 | Stroud H, Ding B, Simon SA, Feng SH, Bellizzi M, Pellegrini M, Wang GL, Meyers BC, Jacobsen SE (2013). Plants regenerated from tissue culture contain stable epigenome changes in rice.eLife 2, e00354. |
| 27 | Thompson DM, Parker R (2009). The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185, 43-50. |
| 28 | Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis.Nat Struct Mol Biol 19, 900-905. |
| 29 | Vare VYP, Eruysal ER, Narendran A, Sarachan KL, Agris PF (2017). Chemical and conformational diversity of modified nucleosides affects tRNA structure and function.Bio- molecules 7, 29. |
| 30 | Wang F, Johnson NR, Coruh C, Axtell MJ (2016a). Genome-wide analysis of single non-templated nucleotides in plant endogenous siRNAs and miRNAs.Nucleic Acids Res 44, 7395-7405. |
| 31 | Wang QH, Li TT, Xu K, Zhang W, Wang XL, Quan JL, Jin WB, Zhang MX, Fan GJ, Wang MB, Shan WX (2016b). The tRNA-derived small RNAs regulate gene expression through triggering sequence-specific degradation of target transcripts in the oomycete pathogen Phytophthora sojae. Front Plant Sci 7, 1938. |
| 32 | Wei LQ, Xu WY, Deng ZY, Su Z, Xue YB, Wang T (2010). Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11, 338. |
| 33 | Wei LQ, Yan LF, Wang T (2011). Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol 12, R53. |
| [1] | Feifei Zhang, Tianfeng Yang, Lirong Chen, Dongmei Liu, Liuyuan Yang, Duyu Yang, Peng Ju, Lu Lu. Review of pollen color diversity in Angiosperms [J]. Biodiv Sci, 2024, 32(1): 23346-. |
| [2] | Bao Zhu, Jiangzhe Zhao, Kewei Zhang, Peng Huang. Oryza sativa CYTOKININ OXIDASE 9(OsCKX9)Is Involved in Regulating the Rice Lamina Joint Development and Leaf Angle [J]. Chinese Bulletin of Botany, 2024, 59(1): 0-0. |
| [3] | Yanli Fang, Chuanyu Tian, Ruyi Su, Yapei Liu, Chunlian Wang, Xifeng Chen, Wei Guo, Zhiyuan Ji. Mining and Preliminary Mapping of Rice Resistance Genes against Bacterial Leaf Streak [J]. Chinese Bulletin of Botany, 2024, 59(1): 0-0. |
| [4] | Tian Chuanyu, Fang Yanli, Shen Qing, Wang Hongjie, Chen Xifeng, Guo Wei, Zhao Kaijun, Wang Chunlian, Ji Zhiyuan. Genotypic Diversity and Pathogenisity of Xanthomonas oryzae pv. oryzae Isolated from Southern China in 2019-2021 [J]. Chinese Bulletin of Botany, 2023, 58(5): 743-749. |
| [5] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [6] | Fan Wu, Shenyun Liu, Huqiang Jiang, Qian Wang, Kaiwei Chen, Hongliang Li. Pollination difference between Apis cerana cerana and Apis mellifera ligustica during the late autumn and winter [J]. Biodiv Sci, 2023, 31(5): 22528-. |
| [7] | Xiongbo Peng, Meng-xiang Sun. Out of the Road: Novel Finding in Regulatory Mechanism of Angiosperm Fertilization [J]. Chinese Bulletin of Botany, 2023, 58(4): 515-518. |
| [8] | Shang Sun, Yingying Hu, Yangshuo Han, Chao Xue, Zhiyun Gong. Double-stranded Labelled Oligo-FISH in Rice Chromosomes [J]. Chinese Bulletin of Botany, 2023, 58(3): 433-439. |
| [9] | Jiayi Jin, Yiting Luo, Huimin Yang, Tao Lu, Hanfei Ye, Jiyi Xie, Kexin Wang, Qianyu Chen, Yuan Fang, Yuexing Wang, Yuchun Rao. QTL Mapping and Expression Analysis on Candidate Genes Related to Chlorophyll Content in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 394-403. |
| [10] | Yuping Yan, Xiaoqi Yu, Deyong Ren, Qian Qian. Genetic Mechanisms and Breeding Utilization of Grain Number Per Panicle in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 359-372. |
| [11] | Yuqiang Liu, Jianmin Wan. The Host Controls the Protein Level of Insect Effectors to Balance Immunity and Growth [J]. Chinese Bulletin of Botany, 2023, 58(3): 353-355. |
| [12] | Jingjing Zhao, Haibin Jia, Tien Ming Lee. Market status and the sustainable utilization strategy of wild earthworm (earth dragon) for medicinal use [J]. Biodiv Sci, 2023, 31(3): 22478-. |
| [13] | Qi Wang, Yunzhe Wu, Xueying Liu, Lili Sun, Hong Liao, Xiangdong Fu. The Rice Receptor-like Kinases Function as Key Regulators of Plant Development and Adaptation to the Environment [J]. Chinese Bulletin of Botany, 2023, 58(2): 199-213. |
| [14] | ZHOU Bo-Rui, LIAO Meng-Na, LI Kai, XU De-Yu, CHEN Hai-Yan, NI Jian, CAO Xian-Yong, KONG Zhao-Chen, XU Qing-Hai, ZHANG Yun, Ulrike HERZSCHUH, CAI Yong-Li, CHEN Bi-Shan, CHEN Jing-An, CHEN Ling-Kang, CHENG Bo, GAO Yang, $\boxed{\hbox{HUANG Ci-Xuan}}$ , HUANG Xiao-Zhong, LI Sheng-Feng, LI Wen-Yi, LIU Kam-Biu, LIU Guang-Xiu, LIU Ping-Mei, LIU Xing-Qi, MA Chun-Mei, SONG Chang-Qing, SUN Xiang-Jun, TANG Ling-Yu, WANG Man-Hua, WANG Yong-Bo, $\boxed{\hbox{XIA Yu-Mei}}$ , XU Jia-Sheng, YAN Shun, YANG Xiang-Dong, YAO Yi-Feng, YE Chuan-Yong, ZHANG Zhi-Yong, ZHAO Zeng-You, ZHENG Zhuo, ZHU Cheng. A fossil pollen dataset of China [J]. Chin J Plant Ecol, 2023, 47(10): 1453-1463. |
| [15] | Liu Xiaolong, Ji Ping, Yang Hongtao, Ding Yongdian, Fu Jialing, Liang Jiangxia, Yu Congcong. Priming Effect of Abscisic Acid on High Temperature Stress During Rice Heading-flowering Stage [J]. Chinese Bulletin of Botany, 2022, 57(5): 596-610. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||