

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (4): 548-559.DOI: 10.11983/CBB22147 cstr: 32102.14.CBB22147
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Rong Sun1, Yulu Yang2, Yajun Li2, Hui Zhang1, Xukai Li1( )
)
Received:2022-07-05
Accepted:2022-10-09
Online:2023-07-01
Published:2022-11-02
Contact:
*E-mail: xukai_li@sxau.edu.cnRong Sun, Yulu Yang, Yajun Li, Hui Zhang, Xukai Li. Genome-wide Identification and Analysis of PLATZ Transcription Factor Gene Family in Foxtail Millet[J]. Chinese Bulletin of Botany, 2023, 58(4): 548-559.
| Gene name | Locus ID | Protein length (aa) | Molecular weight (kDa) | Isoelectric point | Instability index | Predicted sub-cellular localization |
|---|---|---|---|---|---|---|
| SiPLATZ1 | Si1g06060 | 210 | 22.98 | 6.65 | 45.46 | Nucleus |
| SiPLATZ2 | Si1g06070 | 182 | 19.81 | 9.32 | 43.97 | Nucleus |
| SiPLATZ3 | Si1g06080 | 248 | 26.72 | 5.45 | 45.38 | Nucleus, mitochondrion |
| SiPLATZ4 | Si1g06100 | 143 | 15.56 | 9.05 | 42.96 | Nucleus, chloroplast |
| SiPLATZ5 | Si1g06720 | 237 | 27.18 | 8.75 | 54.77 | Nucleus |
| SiPLATZ6 | Si1g07690 | 257 | 27.79 | 8.63 | 45.93 | Chloroplast |
| SiPLATZ7 | Si1g26800 | 198 | 21.94 | 9.05 | 69.15 | Nucleus |
| SiPLATZ8 | Si1g28470 | 247 | 27.37 | 9.07 | 56.56 | Nucleus |
| SiPLATZ9 | Si4g17460 | 242 | 26.07 | 8.65 | 47.57 | Nucleus, cell membrane, chloroplast |
| SiPLATZ10 | Si4g20160 | 243 | 26.84 | 9.47 | 60.37 | Nucleus |
| SiPLATZ11 | Si5g17000 | 214 | 24.38 | 7.90 | 63.39 | Nucleus |
| SiPLATZ12 | Si5g19690 | 209 | 23.92 | 6.50 | 66.21 | Nucleus |
| SiPLATZ13 | Si6g24710 | 207 | 22.35 | 9.15 | 47.83 | Nucleus, chloroplast |
| SiPLATZ14 | Si7g22910 | 251 | 27.42 | 9.23 | 51.67 | Nucleus |
| SiPLATZ15 | Si9g30780 | 246 | 26.90 | 9.16 | 56.51 | Nucleus |
| SiPLATZ16 | Si9g36990 | 256 | 29.23 | 6.02 | 70.27 | Nucleus |
| SiPLATZ17 | Si9g47270 | 239 | 26.34 | 9.37 | 52.33 | Nucleus |
Table 1 Physicochemical properties of the proteins encoded by SiPLATZ gene family in foxtail millet
| Gene name | Locus ID | Protein length (aa) | Molecular weight (kDa) | Isoelectric point | Instability index | Predicted sub-cellular localization |
|---|---|---|---|---|---|---|
| SiPLATZ1 | Si1g06060 | 210 | 22.98 | 6.65 | 45.46 | Nucleus |
| SiPLATZ2 | Si1g06070 | 182 | 19.81 | 9.32 | 43.97 | Nucleus |
| SiPLATZ3 | Si1g06080 | 248 | 26.72 | 5.45 | 45.38 | Nucleus, mitochondrion |
| SiPLATZ4 | Si1g06100 | 143 | 15.56 | 9.05 | 42.96 | Nucleus, chloroplast |
| SiPLATZ5 | Si1g06720 | 237 | 27.18 | 8.75 | 54.77 | Nucleus |
| SiPLATZ6 | Si1g07690 | 257 | 27.79 | 8.63 | 45.93 | Chloroplast |
| SiPLATZ7 | Si1g26800 | 198 | 21.94 | 9.05 | 69.15 | Nucleus |
| SiPLATZ8 | Si1g28470 | 247 | 27.37 | 9.07 | 56.56 | Nucleus |
| SiPLATZ9 | Si4g17460 | 242 | 26.07 | 8.65 | 47.57 | Nucleus, cell membrane, chloroplast |
| SiPLATZ10 | Si4g20160 | 243 | 26.84 | 9.47 | 60.37 | Nucleus |
| SiPLATZ11 | Si5g17000 | 214 | 24.38 | 7.90 | 63.39 | Nucleus |
| SiPLATZ12 | Si5g19690 | 209 | 23.92 | 6.50 | 66.21 | Nucleus |
| SiPLATZ13 | Si6g24710 | 207 | 22.35 | 9.15 | 47.83 | Nucleus, chloroplast |
| SiPLATZ14 | Si7g22910 | 251 | 27.42 | 9.23 | 51.67 | Nucleus |
| SiPLATZ15 | Si9g30780 | 246 | 26.90 | 9.16 | 56.51 | Nucleus |
| SiPLATZ16 | Si9g36990 | 256 | 29.23 | 6.02 | 70.27 | Nucleus |
| SiPLATZ17 | Si9g47270 | 239 | 26.34 | 9.37 | 52.33 | Nucleus |
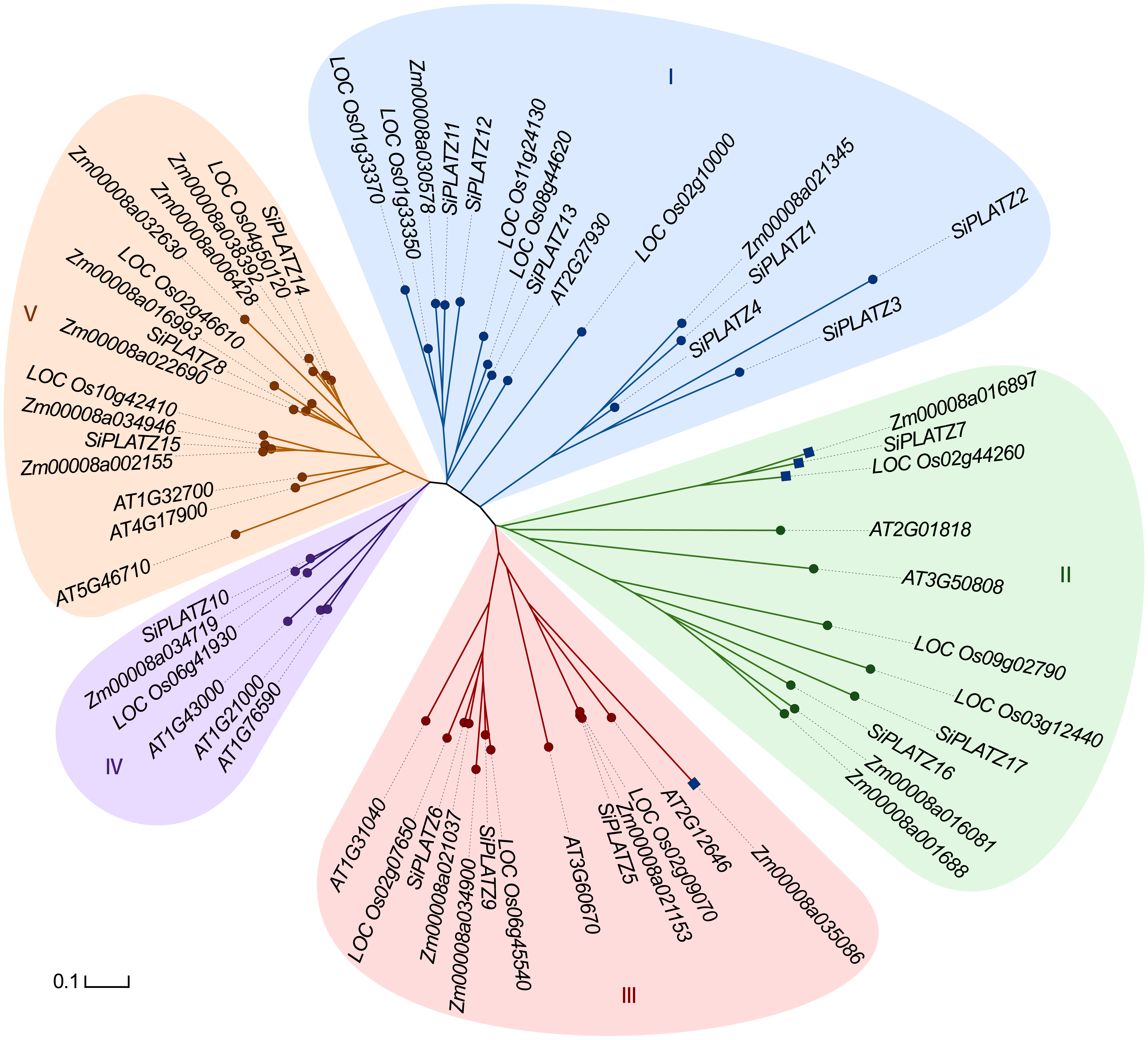
Figure 1 Phylogenetic relationships of PLATZ family proteins in foxtail millet, rice, maize and Arabidopsis The branch color and filled base color on the phylogenetic tree distinguish subgroups of the PLATZ gene family, and the branch length indicates the genetic distance of the gene. Genes in round nodes are consistent with the grouping of Wang et al. (2018), genes in square nodes (4 genes) are different from that, and genes with the same node colors indicate in the same subgroup of the Wang et al. (2018).
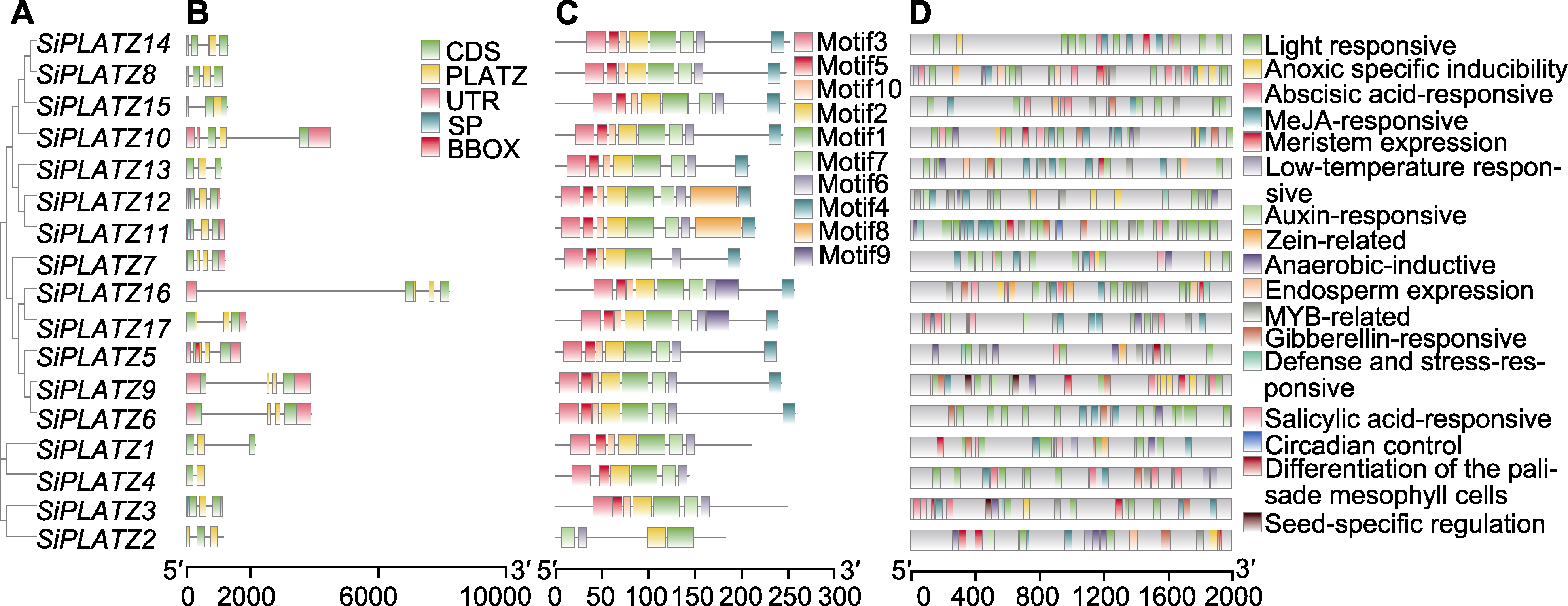
Figure 2 Structural analysis of PLATZ family genes in foxtail millet (A) Evolutionary tree of PLATZ family in foxtail millet; (B) The gene structure of PLATZ family in foxtail millet (CDS: Coding sequences; PLATZ: Plant AT-rich sequence and zinc-binding protein domains; UTR: Untranslated regions; SP: Signal peptide; BBOX: B-Box-type zinc finger domains); (C) The conserved motif of PLATZ family in foxtail millet; (D) Analysis of cis-acting elements in the promoter of PLATZ family gene in foxtail millet
| Homologous gene | Ka | Ks | Ka/Ks |
|---|---|---|---|
| SiPLATZ10-SiPLATZ4 segmental duplication | 0.434473 | 0.577238 | 0.752677 |
| SiPLATZ14-SiPLATZ8 segmental duplication | 0.164238 | 0.394477 | 0.416344 |
| SiPLATZ1-SiPLATZ2 tandem repeats | 0.471405 | 1.482013 | 0.318084 |
| SiPLATZ1-SiPLATZ3 tandem repeats | 0.415490 | 0.958245 | 0.433595 |
| SiPLATZ1-SiPLATZ4 tandem repeats | 0.173058 | 0.550660 | 0.314274 |
| SiPLATZ2-SiPLATZ3 tandem repeats | 0.516098 | 0.872575 | 0.591465 |
| SiPLATZ2-SiPLATZ4 tandem repeats | 0.614008 | 1.027812 | 0.597393 |
| SiPLATZ3-SiPLATZ4 tandem repeats | 0.352090 | 0.772969 | 0.455503 |
Table 2 Calculation of Ka/Ks value of SiPLATZ collinear gene pair in foxtail millet genome by NG method
| Homologous gene | Ka | Ks | Ka/Ks |
|---|---|---|---|
| SiPLATZ10-SiPLATZ4 segmental duplication | 0.434473 | 0.577238 | 0.752677 |
| SiPLATZ14-SiPLATZ8 segmental duplication | 0.164238 | 0.394477 | 0.416344 |
| SiPLATZ1-SiPLATZ2 tandem repeats | 0.471405 | 1.482013 | 0.318084 |
| SiPLATZ1-SiPLATZ3 tandem repeats | 0.415490 | 0.958245 | 0.433595 |
| SiPLATZ1-SiPLATZ4 tandem repeats | 0.173058 | 0.550660 | 0.314274 |
| SiPLATZ2-SiPLATZ3 tandem repeats | 0.516098 | 0.872575 | 0.591465 |
| SiPLATZ2-SiPLATZ4 tandem repeats | 0.614008 | 1.027812 | 0.597393 |
| SiPLATZ3-SiPLATZ4 tandem repeats | 0.352090 | 0.772969 | 0.455503 |
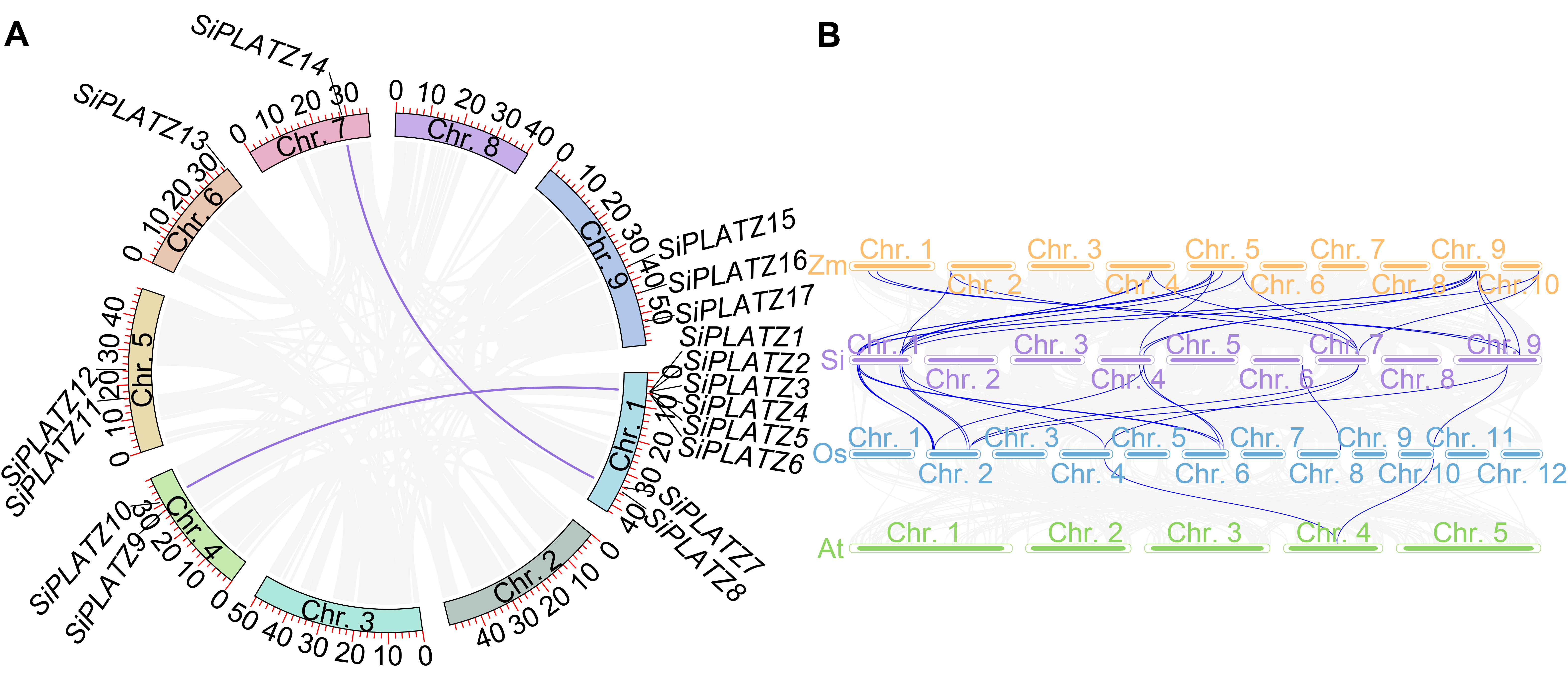
Figure 3 Collinear analysis of PLATZ family genes (A) The internal collinearity of the PLATZ family in foxtail millet; (B) The collinearity of the PLATZ family in maize, foxtail millet, rice and Arabidopsis
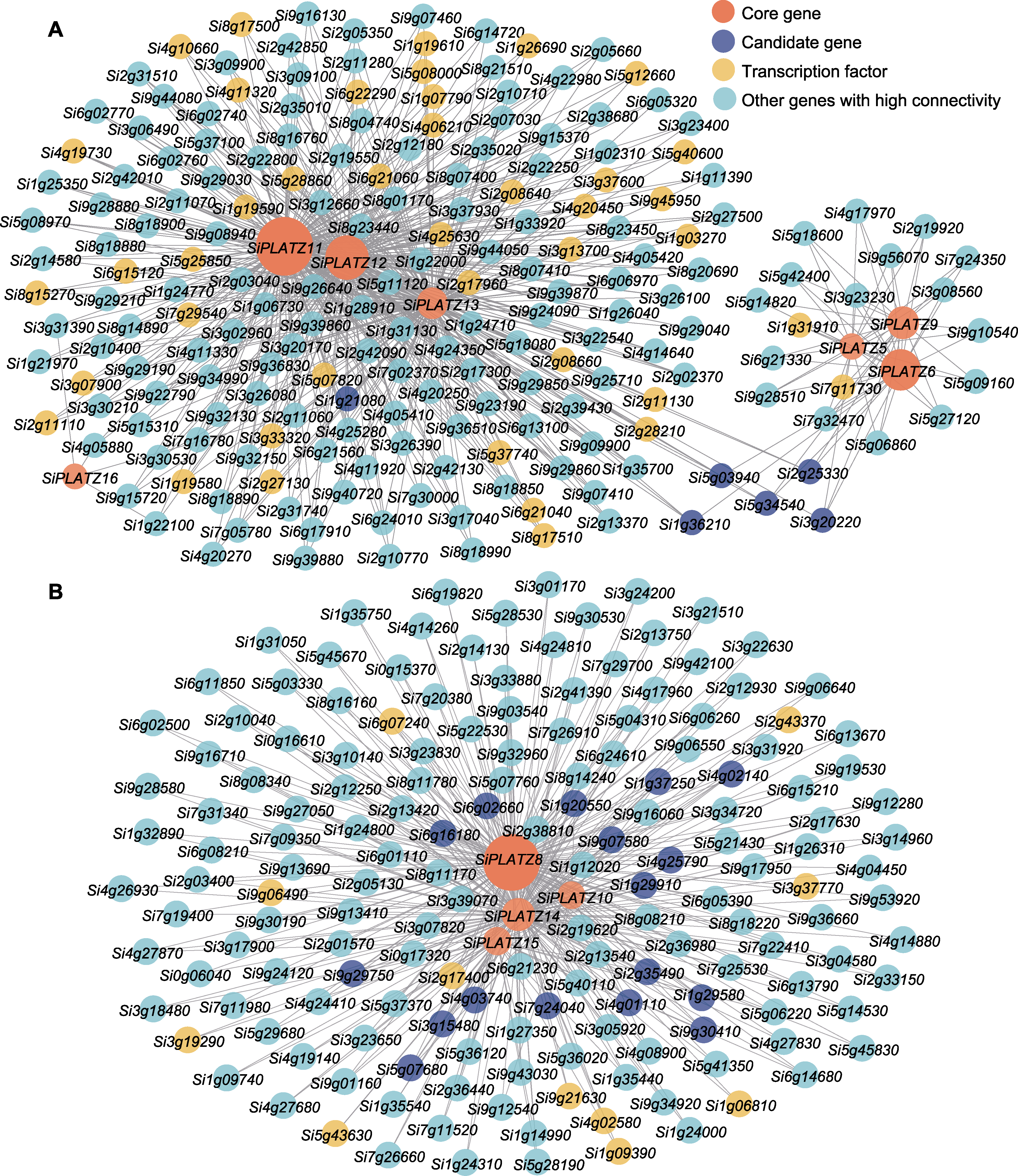
Figure 4 Local network of genes related to the PLATZ family of foxtail millet (A) The related gene network diagram of the foxtail millet PLATZ family in the blue module; (B) The related gene network diagram of the foxtail millet PLATZ family in the turquoise module. Red indicates the core gene, blue indicates the candidate gene, yellow indicates the transcription factor, and green indicates other co-expressed genes with high connectivity. The size of the circle represented the level of gene connectivity. The larger the circle, the higher the connectivity, and vice versa.
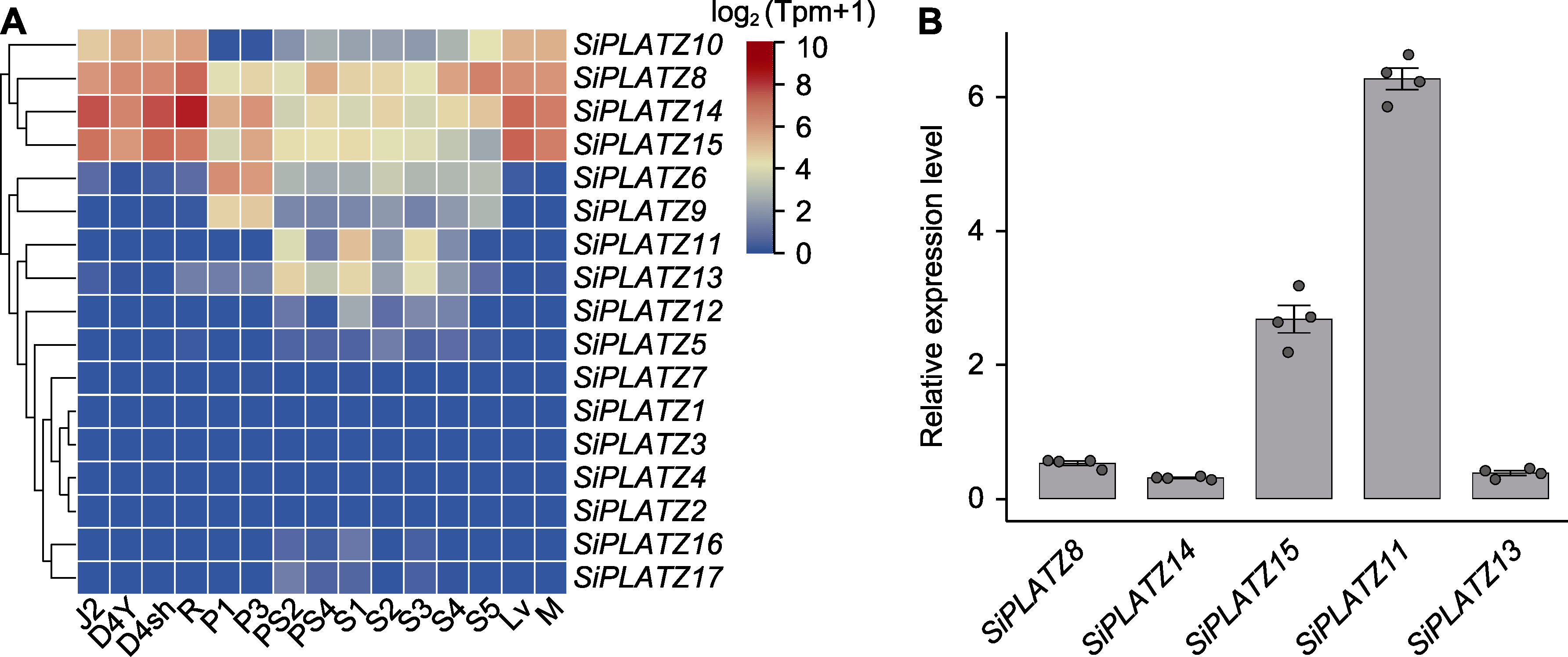
Figure 5 Expression patterns of SiPLATZ family members in different tissue and development stages of foxtail millet (A) The expression levels of SiPLATZ family members in different tissue stages of foxtail millet (it consists of 15 tissue stages, J2: Stem-top-second_filling-stage; D4Y: Leaf-top-fourth_filling-stage; D4sh: Leaf-sheath-top-fourth_filling-stage; R: Root_filling- stage; P1: Panicle_primary-panicle-branch-differentiation-stage; P3: Panicle_third-panicle-branch-differentiation-stage; PS2: Immature-spikelet_S2; PS4: Immature-spikelet_S4; S1-S5: Immature-seed_S1-S5; Lv: Leaf-veins_S3; M: Mesophyll_S3); (B) qPCR verification of part of the SiPLATZ genes.
| [1] | 陈睿, 陈建民, 吴明基, 杨绍华, 胡昌泉 (2019). 水稻OsPLATZ14基因启动子的克隆及表达分析. 福建农业学报 34, 1137-1143. |
| [2] |
刘宝玲, 张莉, 孙岩, 薛金爱, 高昌勇, 苑丽霞, 王计平, 贾小云, 李润植 (2016). 谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析. 植物学报 51, 473-487.
DOI |
| [3] |
王海岗, 贾冠清, 智慧, 温琪汾, 董俊丽, 陈凌, 王君杰, 曹晓宁, 刘思辰, 王纶, 乔治军, 刁现民 (2016). 谷子核心种质表型遗传多样性分析及综合评价. 作物学报 42, 19-30.
DOI |
| [4] | 杨瑞 (2017). 拟南芥锌指转录因子PLATZ5在盐胁迫响应中的功能研究. 硕士论文. 泰安: 山东农业大学. pp. 1-66. |
| [5] | 张亚 (2018). 锌依赖的DNA结合蛋白家族基因OsPLATZ的功能研究. 硕士论文. 广州: 华南农业大学. pp. 1-69. |
| [6] |
赵娟, 尹艺臻, 王晓璐, 马春英, 尹美强, 温银元, 宋喜娥, 董淑琦, 杨雪芳, 原向阳 (2020). 不同品种谷子愈伤组织对拿捕净胁迫的生理响应. 中国农业科学 53, 917-928.
DOI |
| [7] | Bailey TL, Johnson J, Grant CE, Noble WS (2015). The MEME suite. Nucleic Acids Res 43, W39-W49. |
| [8] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13, 1194-1202.
DOI PMID |
| [9] | Chou KC, Shen HB (2010). Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat Sci 2, 1090-1103. |
| [10] | Diao XM, Schnable J, Bennetzen JL, Li JY (2014). Initiation of Setaria as a model plant. Front Agric Sci Eng 1, 16-20. |
| [11] | Duvaud S, Gabella C, Lisacek F, Stockinger H, Ioannidis V, Durinx C (2021). Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res 49, W216-W227. |
| [12] |
Fu YX, Cheng MP, Li ML, Guo XJ, Wu YR, Wang JR (2020). Identification and characterization of PLATZ transcription factors in wheat. Int J Mol Sci 21, 8934.
DOI URL |
| [13] | Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40, D1178-D1186. |
| [14] |
Huangfu YG, Pan JW, Li Z, Wang QG, Mastouri F, Li Y, Yang S, Liu M, Dai SJ, Liu W (2021). Genome-wide identification of PTI1 family in Setaria italica and salinity-responsive functional analysis of SiPTI1-5. BMC Plant Biol 21, 319.
DOI |
| [15] |
Jones MK, Liu XY (2009). Origins of agriculture in East Asia. Science 324, 730-731.
DOI URL |
| [16] |
Kim JH, Kim J, Jun SE, Park S, Timilsina R, Kwon DS, Kim Y, Park SJ, Hwang JY, Nam HG, Kim GT, Woo HR (2018). ORESARA15, a PLATZ transcription factor, mediates leaf growth and senescence in Arabidopsis. New Phytol 220, 609-623.
DOI URL |
| [17] |
Langfelder P, Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559.
DOI PMID |
| [18] |
Lee GA, Crawford GW, Liu L, Chen XC (2007). Plants and people from the Early Neolithic to Shang periods in North China. Proc Natl Acad Sci USA 104, 1087-1092.
DOI URL |
| [19] | Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30, 325-327. |
| [20] | Letunic I, Khedkar S, Bork P (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49, D458-D460. |
| [21] |
Li Q, Wang JC, Ye JW, Zheng XX, Xiang XL, Li CS, Fu MM, Wang Q, Zhang ZY, Wu YR (2017). The maize imprinted gene Floury3 encodes a PLATZ protein required for tRNA and 5S rRNA transcription through interaction with RNA polymerase III. Plant Cell 29, 2661-2675.
DOI URL |
| [22] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402-408.
DOI PMID |
| [23] | Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A (2021). Pfam: the protein families database in 2021. Nucleic Acids Res 49, D412-D419. |
| [24] |
Nagano Y, Furuhashi H, Inaba T, Sasaki Y (2001). A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences. Nucleic Acids Res 29, 4097-4105.
DOI PMID |
| [25] | Robinson JT, Thorvaldsdóttir H, Turner D, Mesirov JP (2020). igv.js: an embeddable JavaScript implementation of the integrative genomics viewer (IGV). BioRxiv. [2020-05-05]. https://doi.org/10.1101/2020.05.03.075499. |
| [26] |
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498-2504.
DOI PMID |
| [27] |
Tamura K, Stecher G, Kumar S (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38, 3022-3027.
DOI PMID |
| [28] |
Wang AH, Hou QQ, Si LZ, Huang XH, Luo JH, Lu DF, Zhu JJ, Shangguan YY, Miao JS, Xie YF, Wang YC, Zhao Q, Feng Q, Zhou CC, Li Y, Fan DL, Lu YQ, Tian QL, Wang ZX, Han B (2019). The PLATZ transcription factor GL6 affects grain length and number in rice. Plant Physiol 180, 2077-2090.
DOI PMID |
| [29] |
Wang J, Li SM, Lan L, Xie MS, Cheng S, Gan XL, Huang G, Du GH, Yu K, Ni XM, Liu BL, Peng GX (2021). De novo genome assembly of a foxtail millet cultivar Huagu11 uncovered the genetic difference to the cultivar Yugu1, and the genetic mechanism of imazethapyr tolerance. BMC Plant Biol 21, 271.
DOI PMID |
| [30] |
Wang JC, Ji C, Li Q, Zhou Y, Wu YR (2018). Genome-wide analysis of the plant-specific PLATZ proteins in maize and identification of their general role in interaction with RNA polymerase III complex. BMC Plant Biol 18, 221.
DOI PMID |
| [31] |
Wu JD, Jiang CP, Zhu HS, Jiang HY, Cheng BJ, Zhu SW (2015). Cloning and functional analysis of the promoter of a maize starch synthase III gene (ZmDULL1). Genet Mol Res 14, 5468-5479.
DOI PMID |
| [32] |
Xu WY, Tang WS, Wang CX, Ge LH, Sun JC, Qi X, He Z, Zhou YB, Chen J, Xu ZS, Ma YZ, Chen M (2020). SiMYB56 confers drought stress tolerance in transgenic rice by regulating lignin biosynthesis and ABA signaling pathway. Front Plant Sci 11, 785.
DOI URL |
| [33] |
Yamada M, Han XW, Benfey PN (2020). RGF1 controls root meristem size through ROS signaling. Nature 577, 85-88.
DOI |
| [34] |
Yang ZR, Zhang HS, Li XK, Shen HM, Gao JH, Hou SY, Zhang B, Mayes S, Bennett M, Ma JX, Wu CY, Sui Y, Han YH, Wang XC (2020). A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat Plants 6, 1167-1178.
DOI |
| [35] |
Zhou SR, Xue HW (2020). The rice PLATZ protein SHORT GRAIN6 determines grain size by regulating spikelet hull cell division. J Integr Plant Biol 62, 847-864.
DOI |
| [1] | Hui Zhang, Hongkai Liang, Hui Zhi, Linlin Zhang, Xianmin Diao, Guanqing Jia. Analyses on the Transcription and Structure Variation of β-carotene Isomerase Gene Family in Foxtail Millet [J]. Chinese Bulletin of Botany, 2023, 58(1): 34-50. |
| [2] | Qi Wang, Yanli Xu, Peng Yan, Haosheng Dong, Wei Zhang, Lin Lu, Zhiqiang Dong. Effects of PAC on Soil Nitrogen Supply and Leaf Antioxidant Properties in Foxtail Millet at Anthesis Stage [J]. Chinese Bulletin of Botany, 2023, 58(1): 90-107. |
| [3] | Haixia Xu, Jing He, Hang Yi, Li Wang. Sex Specific Response Mechanism of Transcriptome in Both Male and Female Marchantia polymorpha Under Cadmium Stress [J]. Chinese Bulletin of Botany, 2022, 57(2): 182-196. |
| [4] | Lan Yang, Ya Liu, Yang Xiang, Xiujuan Sun, Jingwei Yan, Aying Zhang. Establishment and Optimization of a Shoot Tip-based Genetic Transformation System for Foxtail Millet [J]. Chinese Bulletin of Botany, 2021, 56(1): 71-79. |
| [5] | Baoling Liu, Li Zhang, Yan Sun,Jinai Xue, Changyong Gao, Lixia Yuan, Jiping Wang, Xiaoyun Jia, Runzhi Li. Genome-wide Characterization of bZIP Transcription Factors in Foxtail Millet and Their Expression Profiles in Response to Drought and Salt Stresses [J]. Chinese Bulletin of Botany, 2016, 51(4): 473-487. |
| [6] | Keliang Zhao;Xiaoqiang Li;Xue Shang;Xinying Zhou;Nan Sun;. Agricultural Characteristics of Middle-late Bronze Age in Western Liaoning Province [J]. Chinese Bulletin of Botany, 2009, 44(06): 718-724. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||