

Chinese Bulletin of Botany ›› 2022, Vol. 57 ›› Issue (2): 182-196.DOI: 10.11983/CBB21189 cstr: 32102.14.CBB21189
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Haixia Xu, Jing He, Hang Yi, Li Wang( )
)
Received:2021-11-08
Accepted:2022-02-25
Online:2022-03-01
Published:2022-03-24
Contact:
Li Wang
Haixia Xu, Jing He, Hang Yi, Li Wang. Sex Specific Response Mechanism of Transcriptome in Both Male and Female Marchantia polymorpha Under Cadmium Stress[J]. Chinese Bulletin of Botany, 2022, 57(2): 182-196.
| Gene names | Primer sequences (5'-3') |
|---|---|
| MpPCS | F: GAGTCCTTTCGAGCGAATTCT |
| R: AATCTTCCAGCAGTAGCAGTAG | |
| MpActin | F: AGGCATCTGGTATCCACGAG |
| R: ACATGGTCGTTCCTCCAGAC | |
| MARPO_0086s0070 | F: ATTGTCACTTCAAACTGCTTCC R: CTTTGCCAGTATGCTTTCAACT |
| MARPO_0265s0002 | F: TGGCGACTCTTAAAATAGCTCT R: GAAAGTATGGGATTCTTGCGAC |
| MARPO_0001s0061 | F: GAGGAAAATCAACGATACAGGC R: CTTCTCTTGCAGTCAGTTGTTC |
| MARPO_0145s0031 | F: CGTAGGGAGATCAGTGATTACC R: CCTTGTATAACTCACGATCGGA |
| MARPO_0003s0122 | F: TGGACAGAAAACTCATTGGTCT R: ACTTCCTTCTCTCTCACCAATG |
| MARPO_0003s0268 | F: GACTAGGGTTCCTTGGTTCTAC R: CATAATCGGGAAAACGTAACCC |
| MARPO_0011s0046 | F: CGGCTCAGATGTTGATAAATCA R: ATGCACTTCGCTGATCTCATAA |
| MARPO_0032s0085 | F: CCCCTCATCTCAAACTACTTGT R: TTGGAAAAGGAATTGCTGGATG |
Table 1 Primer sequences for qRT-PCR
| Gene names | Primer sequences (5'-3') |
|---|---|
| MpPCS | F: GAGTCCTTTCGAGCGAATTCT |
| R: AATCTTCCAGCAGTAGCAGTAG | |
| MpActin | F: AGGCATCTGGTATCCACGAG |
| R: ACATGGTCGTTCCTCCAGAC | |
| MARPO_0086s0070 | F: ATTGTCACTTCAAACTGCTTCC R: CTTTGCCAGTATGCTTTCAACT |
| MARPO_0265s0002 | F: TGGCGACTCTTAAAATAGCTCT R: GAAAGTATGGGATTCTTGCGAC |
| MARPO_0001s0061 | F: GAGGAAAATCAACGATACAGGC R: CTTCTCTTGCAGTCAGTTGTTC |
| MARPO_0145s0031 | F: CGTAGGGAGATCAGTGATTACC R: CCTTGTATAACTCACGATCGGA |
| MARPO_0003s0122 | F: TGGACAGAAAACTCATTGGTCT R: ACTTCCTTCTCTCTCACCAATG |
| MARPO_0003s0268 | F: GACTAGGGTTCCTTGGTTCTAC R: CATAATCGGGAAAACGTAACCC |
| MARPO_0011s0046 | F: CGGCTCAGATGTTGATAAATCA R: ATGCACTTCGCTGATCTCATAA |
| MARPO_0032s0085 | F: CCCCTCATCTCAAACTACTTGT R: TTGGAAAAGGAATTGCTGGATG |
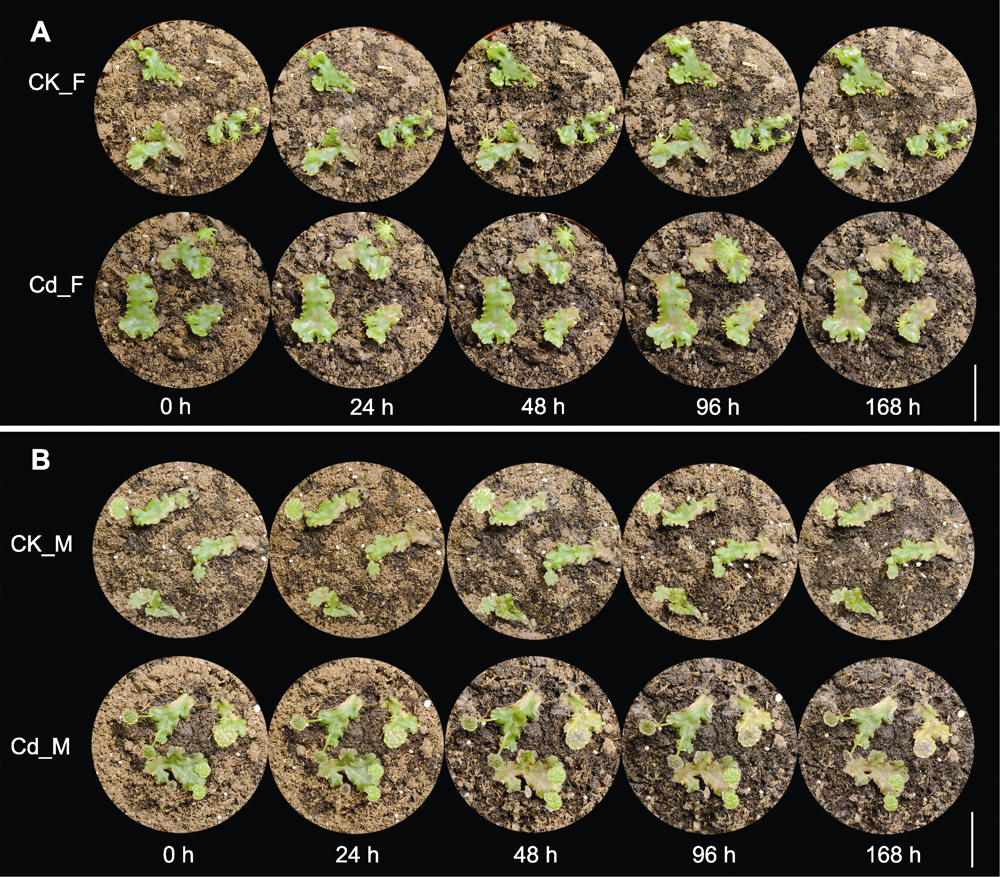
Figure 1 Morphology of female (A) and male (B) Marchantia polymorpha after cadmium (Cd) treatment Five time points after Cd treatment: 0, 24, 48, 96 and 168 h. Bars=2 cm
| Sample_ID | Raw reads | Clean reads | Error (%) | Q20 (%) | Q30 (%) | GC (%) | Total mapped (%) |
|---|---|---|---|---|---|---|---|
| CK_F1 | 50740902 | 50191272 | 0.0241 | 98.38 | 95.02 | 50.92 | 94.74 |
| CK_F2 | 58049938 | 57506790 | 0.0242 | 98.36 | 94.95 | 50.98 | 89.75 |
| CK_F3 | 53602814 | 53058498 | 0.0245 | 98.23 | 94.63 | 51.23 | 95.59 |
| CK_M1 | 49826850 | 49313610 | 0.0243 | 98.31 | 94.84 | 51.14 | 94.22 |
| CK_M2 | 52605578 | 51996578 | 0.0245 | 98.25 | 94.67 | 51.13 | 91.95 |
| CK_M3 | 50064978 | 49523028 | 0.0242 | 98.36 | 94.96 | 50.94 | 90.51 |
| Cd_F1 | 53604084 | 53084914 | 0.0241 | 98.40 | 95.03 | 51.02 | 94.65 |
| Cd_F2 | 48343374 | 47896356 | 0.0255 | 97.87 | 93.60 | 50.53 | 95.16 |
| Cd_F3 | 48556392 | 48058072 | 0.0247 | 98.15 | 94.43 | 50.90 | 96.26 |
| Cd_M1 | 51066022 | 50567610 | 0.0244 | 98.28 | 94.75 | 50.66 | 87.88 |
| Cd_M2 | 51079094 | 50542790 | 0.0244 | 98.28 | 94.75 | 50.48 | 90.25 |
| Cd_M3 | 54825492 | 54241196 | 0.0249 | 98.06 | 94.19 | 50.61 | 93.78 |
Table 2 Transcriptome quality control data
| Sample_ID | Raw reads | Clean reads | Error (%) | Q20 (%) | Q30 (%) | GC (%) | Total mapped (%) |
|---|---|---|---|---|---|---|---|
| CK_F1 | 50740902 | 50191272 | 0.0241 | 98.38 | 95.02 | 50.92 | 94.74 |
| CK_F2 | 58049938 | 57506790 | 0.0242 | 98.36 | 94.95 | 50.98 | 89.75 |
| CK_F3 | 53602814 | 53058498 | 0.0245 | 98.23 | 94.63 | 51.23 | 95.59 |
| CK_M1 | 49826850 | 49313610 | 0.0243 | 98.31 | 94.84 | 51.14 | 94.22 |
| CK_M2 | 52605578 | 51996578 | 0.0245 | 98.25 | 94.67 | 51.13 | 91.95 |
| CK_M3 | 50064978 | 49523028 | 0.0242 | 98.36 | 94.96 | 50.94 | 90.51 |
| Cd_F1 | 53604084 | 53084914 | 0.0241 | 98.40 | 95.03 | 51.02 | 94.65 |
| Cd_F2 | 48343374 | 47896356 | 0.0255 | 97.87 | 93.60 | 50.53 | 95.16 |
| Cd_F3 | 48556392 | 48058072 | 0.0247 | 98.15 | 94.43 | 50.90 | 96.26 |
| Cd_M1 | 51066022 | 50567610 | 0.0244 | 98.28 | 94.75 | 50.66 | 87.88 |
| Cd_M2 | 51079094 | 50542790 | 0.0244 | 98.28 | 94.75 | 50.48 | 90.25 |
| Cd_M3 | 54825492 | 54241196 | 0.0249 | 98.06 | 94.19 | 50.61 | 93.78 |
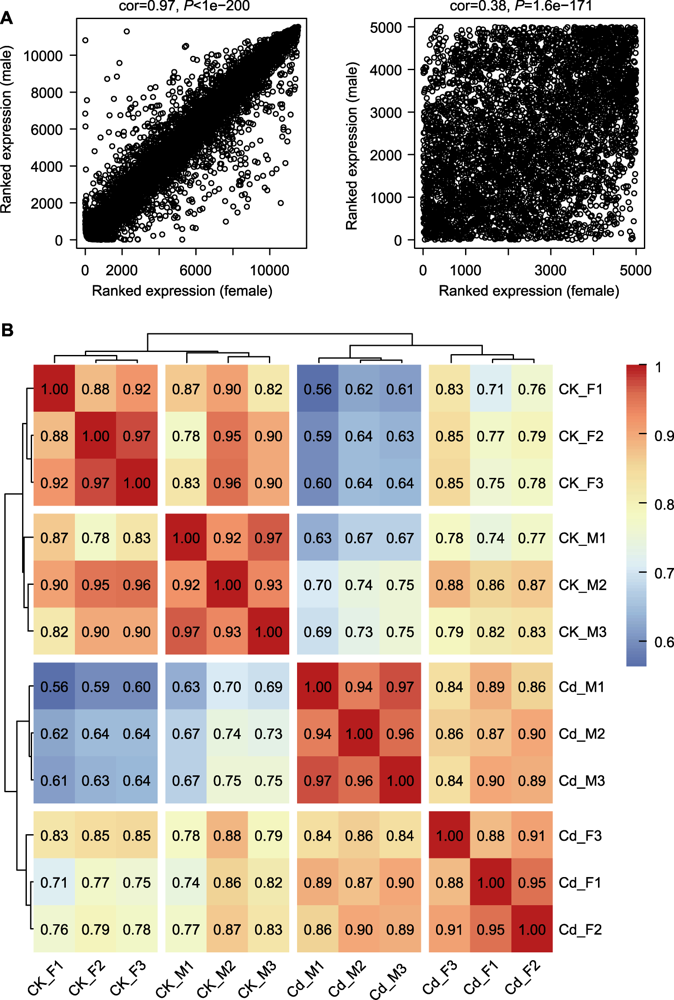
Figure 2 Correlation analysis among female and male Marchantia polymorpha samples (A) Scatter plot of correlation about average level of gene expression and overall connectivity between female and male samples (females as x-axis and males as y-axis); (B) Clustered heatmap of correlation among samples
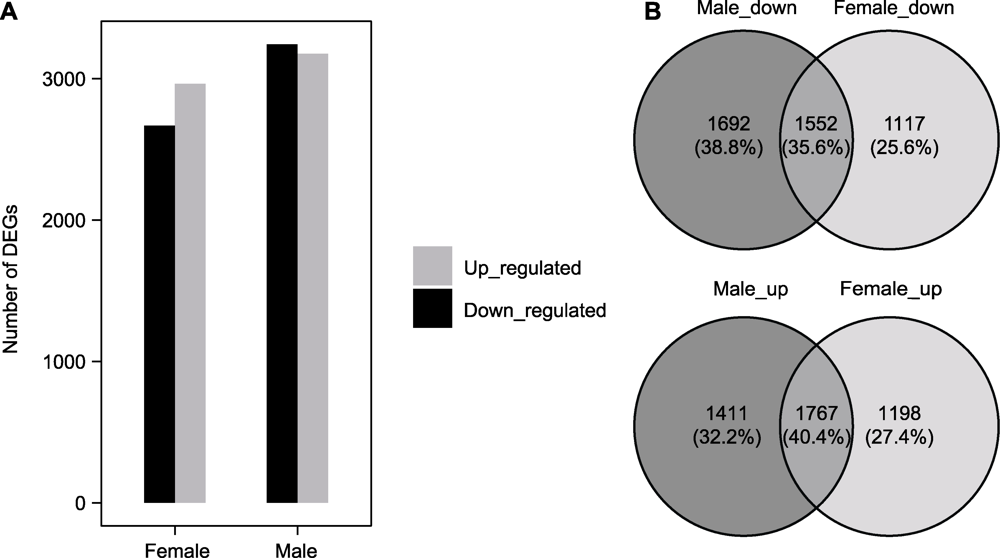
Figure 3 Statistics of differentially expressed genes between female and male Marchantia polymorpha in response to cadmium stress (A) Histograms of differentially expressed genes (DEGs); (B) Venn diagram of number of differential genes with different changing trends
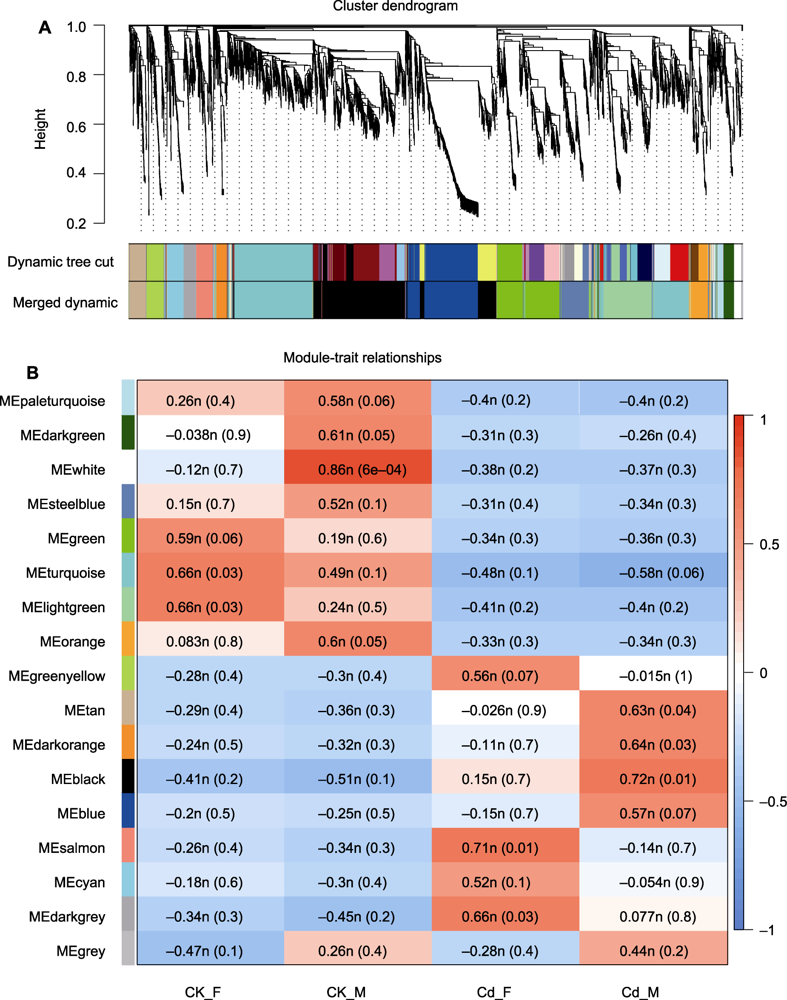
Figure 5 Weighted gene co-expression network analysis of female and male Marchantia polymorpha response to cadmium stress (A) Cluster dendrogram of genes and modules; (B) Heatmap of correlation between modules and phenotypes. Red indicates positive correlation and blue indicates negative correlation.
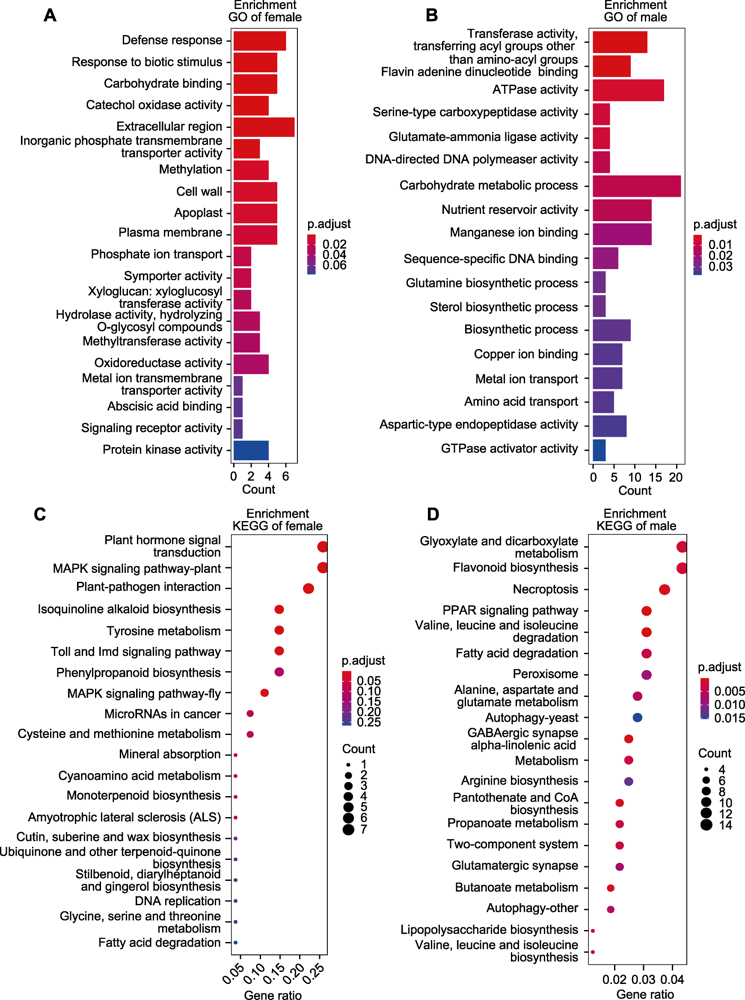
Figure 6 Enrichment analysis of target modules (A) Top ten GO enrichment of salmon module; (B) Top ten GO enrichment of black module; (C) Top ten KEGG enrichment of salmon module; (D) Top ten KEGG enrichment of black module
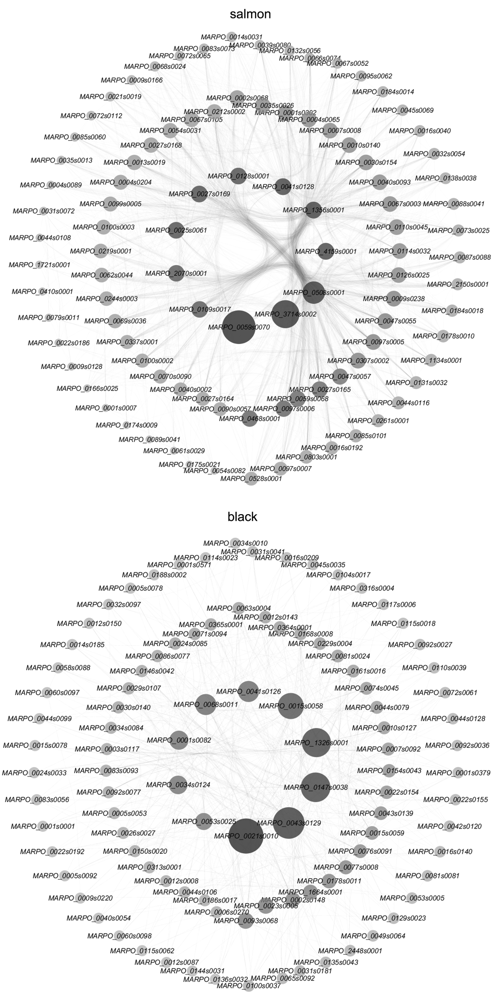
Figure 7 Co-expression network diagram and hub genes of the salmon module and black module The diameter of dot from small to large, with light to dark colors represents the increasement in degree.
| Module | Gene names | Protein names |
|---|---|---|
| black | MARPO_0147s0038 | Phytocyanin domain-containing protein |
| black | MARPO_1326s0001 | ANK_REP_REGION domain-containing protein |
| black | MARPO_0015s0058 | NAC domain-containing protein |
| black | MARPO_0068s0011 | Hydrolase_4 domain-containing protein |
| black | MARPO_0034s0124 | Protein kinase domain-containing protein |
| salmon | MARPO_4159s0001 | Protein kinase domain-containing protein (fragment) |
| salmon | MARPO_0097s0006 | SCP domain-containing protein |
| salmon | MARPO_0047s0057 | FAD-binding FR-type domain-containing protein |
| salmon | MARPO_0009s0238 | S-protein homolog |
| salmon | MARPO_0047s0055 | FAD-binding FR-type domain-containing protein |
Table 3 Functional annotation of hub genes in salmon module and black module
| Module | Gene names | Protein names |
|---|---|---|
| black | MARPO_0147s0038 | Phytocyanin domain-containing protein |
| black | MARPO_1326s0001 | ANK_REP_REGION domain-containing protein |
| black | MARPO_0015s0058 | NAC domain-containing protein |
| black | MARPO_0068s0011 | Hydrolase_4 domain-containing protein |
| black | MARPO_0034s0124 | Protein kinase domain-containing protein |
| salmon | MARPO_4159s0001 | Protein kinase domain-containing protein (fragment) |
| salmon | MARPO_0097s0006 | SCP domain-containing protein |
| salmon | MARPO_0047s0057 | FAD-binding FR-type domain-containing protein |
| salmon | MARPO_0009s0238 | S-protein homolog |
| salmon | MARPO_0047s0055 | FAD-binding FR-type domain-containing protein |
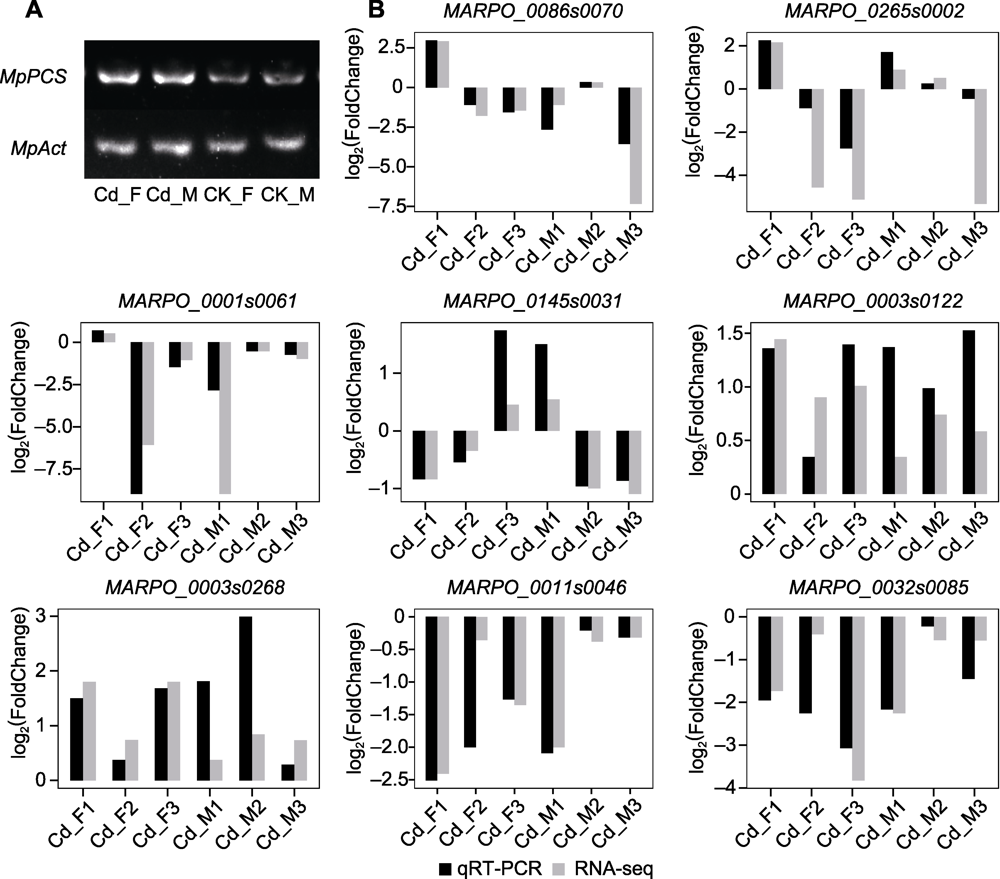
Figure 8 Semi-quantitative analysis of gene expression and expression pattern validation (A) Semi-quantitative RT-PCR was used to detect transcription of marker genes (MpPCS) in four samples, 33 PCR cycles for MpPCS, 27 PCR cycles for MpAct (as an internal reference gene); (B) The expression data were verified by qRT-PCR
| [1] | 安婷婷, 黄帝, 王浩, 张一, 陈应龙 (2021). 植物响应镉胁迫的生理生化机制研究进展. 植物学报 56, 347-362. |
| [2] | 王研, 贾博为, 孙明哲, 孙晓丽 (2021). 野生大豆耐逆分子调控机制研究进展. 植物学报 56, 104-115. |
| [3] |
Arai H, Yanagiura K, Toyama Y, Morohashi K (2019). Genome-wide analysis of MpBHLH12, a IIIf basic helix-loop-helix transcription factor of Marchantia polymorpha. J Plant Res 132, 197-209.
DOI URL |
| [4] |
Ares Á, Itouga M, Kato Y, Sakakibara H (2018). Differential metal tolerance and accumulation patterns of Cd, Cu, Pb and Zn in the liverwort Marchantia polymorpha L. Bull Environ Contam Toxicol 100, 444-450.
DOI URL |
| [5] |
Baruah S, Bora MS, Sharma P, Deb P, Sarma KP (2017). Understanding of the distribution, translocation, bioaccumulation, and ultrastructural changes of Monochoria hastata plant exposed to cadmium. Water Air Soil Pollution 228, 17.
DOI URL |
| [6] |
Bisang I, Hedenäs L (2005). Sex ratio patterns in dioicous bryophytes re-visited. J Bryol 27, 207-219.
DOI URL |
| [7] |
Brown RC, Lemmon BE, Shimamura M, Villarreal JC, Renzaglia KS (2015). Spores of relictual bryophytes: diverse adaptations to life on land. Rev Palaeobot Palynol 216, 1-17.
DOI URL |
| [8] |
Degola F, De Benedictis M, Petraglia A, Massimi A, Fattorini L, Sorbo S, Basile A, di Toppi LS(2014). A Cd/Fe/Zn-responsive phytochelatin synthase is constitutively present in the ancient liverwort Lunularia cruciata (L.) Dumort. Plant Cell Physiol 55, 1884-1891.
DOI URL |
| [9] | Elzanati O, Mouzeyar S, Roche J (2020). Dynamics of the transcriptome response to heat in the moss, Physcomitrella patens. Int J Mol Sci 21, 1512. |
| [10] | Fan CH, Gao Yl, Du B (2016). Response of FTIR and Raman spectra on cell wall of calendula of ficinalis seedlings roots to the co-contamination stress of lead and cadmium in loess. Spectrosc Spect Anal 36, 2076-2081. |
| [11] | Jiao Y, Ge W, Qin R, Sun BL, Jiang WS, Liu DH (2012). Influence of cadmium stress on growth, ultrastructure and antioxidative enzymes in Populus 2025. Fresenius Environ Bull 21, 1375-1384. |
| [12] | Kazemitabar SK, Faraji S, Najafi-Zarrini H (2020). Identification and in silico evaluation of bHLH genes in the Sesamum indicum genome: growth regulation and stress dealing specially through the metal ions homeostasis and flavonoid biosynthesis. Gene Rep 19, 100639. |
| [13] |
Li J, Lu HL, Liu JC, Hong HL, Yan CL (2015). The influence of flavonoid amendment on the absorption of cadmium in Avicennia marina roots. Ecotoxicol Environ Saf 120, 1-6.
DOI URL |
| [14] |
Li JY, Zhang XC, Li D, Sun MY, Shi L (2020a). Energy response patterns to light spectrum at sex differentiation stages of Drynaria roosii gametophytes. Environ Exp Bot 172, 103996.
DOI URL |
| [15] |
Li MG, Barbaro E, Bellini E, Saba A, di Toppi LS, Varotto C (2020b). Ancestral function of the phytochelatin synthase C-terminal domain in inhibition of heavy metal-mediated enzyme overactivation. J Exp Bot 71, 6655-6669.
DOI URL |
| [16] |
Ligrone R, Duckett JG, Renzaglia KS (2012). Major transitions in the evolution of early land plants: a bryological perspective. Ann Bot 109, 851-871.
DOI URL |
| [17] | Liu YK, Liu LX, Qi JH, Dang PY, Xia TS (2019). Cadmium activates ZmMPK3-1 and ZmMPK6-1 via induction of reactive oxygen species in maize roots. Biochem Biophys Res Commun 516, 747-752. |
| [18] |
Lo JC, Tsednee M, Lo YC, Yang SC, Hu JM, Ishizaki K, Kohchi T, Lee DC, Yeh KC (2016). Evolutionary analysis of iron (Fe) acquisition system in Marchantia polymorpha. New Phytol 211, 569-583.
DOI URL |
| [19] |
Markert B, Weckert V (1993). Time-and-site integrated long-term biomonitoring of chemical elements by means of mosses. Toxicol Environ Chem 40, 43-56.
DOI URL |
| [20] |
Marks RA, Smith JJ, Cronk Q, Grassa CJ, McLetchie DN (2019). Genome of the tropical plant Marchantia inflexa: implications for sex chromosome evolution and dehydration tolerance. Sci Rep 9, 8722.
DOI PMID |
| [21] |
Muhammad T, Zhang J, Ma YL, Li YS, Zhang F, Zhang Y, Liang Y (2019). Overexpression of a mitogen-activated protein kinase SlMAPK3 positively regulates tomato tolerance to cadmium and drought stress. Molecules 24, 556.
DOI URL |
| [22] |
Paasch AE, Mishler BD, Nosratinia S, Stark LR, Fisher KM (2015). Decoupling of sexual reproduction and genetic diversity in the female-biased mojave desert moss Syntrichia caninervis (Pottiaceae). Int J Plant Sci 176, 751-761.
DOI URL |
| [23] |
Piper PW, Truman AW, Millson SH, Nuttall J (2006). Hsp90 chaperone control over transcriptional regulation by the yeast Slt2(Mpk1)p and human ERK5 mitogen-activated protein kinases (MAPKs). Biochem Soc Trans 34, 783-785.
DOI URL |
| [24] |
Renner SS, Ricklefs RE (1995). Dioecy and its correlates in the flowering plants. Am J Bot 82, 596-606.
DOI URL |
| [25] | Rio DC, Ares M Jr, Hannon GJ, Nilsen TW (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010, pdb.prot5439. |
| [26] |
Rottenberg A (1998). Sex ratio and gender stability in the dioecious plants of Israel. Bot J Linn Soc 128, 137-148.
DOI URL |
| [27] |
Rubinstein CV, Gerrienne P, de la Puente GS, Astini RA, Steemans P (2010). Early Middle Ordovician evidence for land plants in Argentina (eastern Gondwana). New Phytol 188, 365-369.
DOI PMID |
| [28] |
Siedlecka A, BaszyńAski T (1993). Inhibition of electron flow around photosystem I in chloroplasts of Cd-treated maize plants is due to Cd-induced iron deficiency. Physiol Plant 87, 199-202.
DOI URL |
| [29] |
Sierocka I, Alaba S, Jarmolowski A, Karlowski WM, Szweykowska-Kulinska Z (2020). The identification of differentially expressed genes in male and female gametophytes of simple thalloid liverwort Pellia endiviifolia sp. B using an RNA-seq approach. Planta 252, 21.
DOI PMID |
| [30] |
Sigfridsson KGV, Bernát G, Mamedov F, Styring S (2004). Molecular interference of Cd2+ with photosystem II. Biochim Biophys Acta 1659, 19-31.
PMID |
| [31] |
Thamm A, Saunders TE, Dolan L (2020). MpFEW RHIZOIDS 1 miRNA-mediated lateral inhibition controls rhizoid cell patterning in Marchantia polymorpha. Curr Biol 30, 1905-1915.
DOI PMID |
| [32] |
Wan LC, Zhang HY (2012). Cadmium toxicity: effects on cytoskeleton, vesicular trafficking and cell wall construction. Plant Signal Behav 7, 345-348.
DOI URL |
| [33] |
Wang JX, Zhang XQ, Shi ML, Gao LJ, Niu XF, Te RG, Chen L, Zhang WW (2014). Metabolomic analysis of the salt-sensitive mutants reveals changes in amino acid and fatty acid composition important to long-term salt stress in Synechocystis sp. PCC 6803. Funct Integr Genomics 14, 431-440.
DOI URL |
| [34] |
Wang Q, Zeng XN, Song QL, Sun Y, Feng YJ, Lai YC (2020). Identification of key genes and modules in response to cadmium stress in different rice varieties and stem nodes by weighted gene co-expression network analysis. Sci Rep 10, 9525.
DOI PMID |
| [35] |
Wu MX, Luo Q, Zhao Y, Long Y, Liu SL, Pan YZ (2018). Physiological and biochemical mechanisms preventing Cd toxicity in the new hyperaccumulator Abelmoschus manihot. J Plant Growth Regul 37, 709-718.
DOI URL |
| [36] |
Xin XY, Chen WH, Wang B, Zhu F, Li Y, Yang HL, Li JG, Ren DT (2018). Arabidopsis MKK10-MPK6 mediates red- light-regulated opening of seedling cotyledons through phosphorylation of PIF3. J Exp Bot 69, 423-439.
DOI URL |
| [37] |
Zhang R, Liu JY, Liu QS, He HG, Xu X, Dong TF (2019). Sexual differences in growth and defence of Populus yunnanensis under drought stress. Can J Forest Res 49, 491-499.
DOI |
| [38] | Zhang ZQ, Hu XN, Zhang YQ, Miao ZY, Xie C, Meng XZ, Deng J, Wen JQ, Mysore KS, Frugier F, Wang T, Dong JL (2016). Opposing control by transcription factors MYB61 and MYB3 increases freezing tolerance by relieving C-repeat binding factor suppression. Plant Physiol 172, 1306-1323. |
| [39] |
Zvereva EL, Kozlov MV (2011). Impacts of industrial polluters on bryophytes: a meta-analysis of observational studies. Water Air Soil Pollut 218, 573-586.
DOI URL |
| [1] | Tong Miao, Wang Huan, Zhang Wenshuang, Wang Chao, Song Jianxiao. Distribution characteristics of antibiotic resistance genes in soil bacterial communities exposed to heavy metal pollution [J]. Biodiv Sci, 2025, 33(3): 24101-. |
| [2] | Yaping Wang, Wenquan Bao, Yu’e Bai. Advances in the Application of Single-cell Transcriptomics in Plant Growth, Development and Stress Response [J]. Chinese Bulletin of Botany, 2025, 60(1): 101-113. |
| [3] | Chunjiao Xia, Yunguang Li, Shu Xia, Wei Pang, Chunli Chen. Flow Cytometric Analysis and Sorting in Plant Genomics [J]. Chinese Bulletin of Botany, 2024, 59(5): 774-782. |
| [4] | Yaqi Zhang, Fuxi Rong, Yuxin Shen, Zheyuan Hong, Lantian Zhang, Liang Wu. Research Advances of Structure and Function of HIPP Family in Plants [J]. Chinese Bulletin of Botany, 2024, 59(4): 659-670. |
| [5] | Cai Shuyu, Liu Jianxin, Wang Guofu, Wu Liyuan, Song Jiangping. Regulatory Mechanism of Melatonin on Tomato Seed Germination Under Cd2+ Stress [J]. Chinese Bulletin of Botany, 2023, 58(5): 720-732. |
| [6] | BAI Xue, LI Yu-Jing, JING Xiu-Qing, ZHAO Xiao-Dong, CHANG Sha-Sha, JING Tao-Yu, LIU Jin-Ru, ZHAO Peng-Yu. Response mechanisms of millet and its rhizosphere soil microbial communities to chromium stress [J]. Chin J Plant Ecol, 2023, 47(3): 418-433. |
| [7] | Siyao Liu, Zhu Li, Xin Ke, Lina Sun, Longhua Wu, Jiejie Zhao. Community characteristics of soil collembola around a typical mercury-thallium mining area in Guizhou Province [J]. Biodiv Sci, 2022, 30(12): 22265-. |
| [8] | Yanan Zhang, Lei Huang, Jiabin Li, Lei Zhang, Zhenhua Dang. Identification and Development of Polymorphic Genic-SSRs in Tamarix ramosissima in Alxa Region Based on Transcriptome [J]. Chinese Bulletin of Botany, 2021, 56(4): 433-442. |
| [9] | Run Liu, Zhaohui Zhang, Jiachen Shen, Zhihui Wang. Community characteristics of bryophyte in Karst caves and its effect on heavy metal pollution: A case study of Zhijin Cave, Guizhou Province [J]. Biodiv Sci, 2018, 26(12): 1277-1288. |
| [10] | Quan CHEN, Ke-Ming MA. Effects of Spartina alterniflora invasion on enrichment of sedimental heavy metals in a mangrove wetland and the underlying mechanisms [J]. Chin J Plant Ecol, 2017, 41(4): 409-417. |
| [11] | Yanyan Lü, Sanxiong Fu, Song Chen, Wei Zhang, Cunkou Qi. RNA-sequencing Analysis of Differentially Expressed Genes in Wild-type and BnERF-transgenic Arabidopsis Under Submergence Treatment [J]. Chinese Bulletin of Botany, 2015, 50(3): 321-330. |
| [12] | Zili Wu, Mengyao Yu, Lu Chen, Jing Wei, Xiaoqin Wang, Yong Hu, Yan Yan, Ping Wan. Transcriptome Analysis of Physcomitrella patens Response to Cadmium Stress by Bayesian Network [J]. Chinese Bulletin of Botany, 2015, 50(2): 171-179. |
| [13] | Qi Li, Xia Ji, Enhui Wang, Hanmei Gao, Yanjun Yi. Using Bryophytes as Biomonitor Atmospheric Heavy Metal Deposition in the City of Qingdao [J]. Chinese Bulletin of Botany, 2014, 49(5): 569-577. |
| [14] | Yuanfeng Cai,Zhongjun Jia. Progress in environmental transcriptomics based on next-generation high-throughput sequencing [J]. Biodiv Sci, 2013, 21(4): 401-410. |
| [15] | Zhiqin Wei, Zhiyong Chen, Rong Qin, Yutao Wang, Shaoshan Li. Cu2+ Induced Local Toxicity and DNA Damage, Cell Death in Roots of Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2013, 48(3): 303-312. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||