

Chinese Bulletin of Botany ›› 2021, Vol. 56 ›› Issue (4): 433-442.DOI: 10.11983/CBB20185 cstr: 32102.14.CBB20185
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Yanan Zhang1, Lei Huang1, Jiabin Li1, Lei Zhang2,3, Zhenhua Dang1,*( )
)
Received:2020-11-20
Accepted:2021-05-27
Online:2021-07-01
Published:2021-06-30
Contact:
Zhenhua Dang
Yanan Zhang, Lei Huang, Jiabin Li, Lei Zhang, Zhenhua Dang. Identification and Development of Polymorphic Genic-SSRs in Tamarix ramosissima in Alxa Region Based on Transcriptome[J]. Chinese Bulletin of Botany, 2021, 56(4): 433-442.
| Population | Longitude (E) | Latitude (N) | Altitude (m) | Habitats |
|---|---|---|---|---|
| TR1 | 101°00′06″ | 41°52′16″ | 939 | Wetland |
| TR2 | 101°11′44″ | 41°57′27″ | 927 | Sandy land |
| TR3 | 101°03′36″ | 42°07′02″ | 915 | Wetland |
| TR4 | 101°12′26″ | 41°58′45″ | 924 | Wetland |
| TR5 | 101°16′25″ | 42°01′55″ | 921 | Wetland |
Table 1 Detailed information of the sampling locations
| Population | Longitude (E) | Latitude (N) | Altitude (m) | Habitats |
|---|---|---|---|---|
| TR1 | 101°00′06″ | 41°52′16″ | 939 | Wetland |
| TR2 | 101°11′44″ | 41°57′27″ | 927 | Sandy land |
| TR3 | 101°03′36″ | 42°07′02″ | 915 | Wetland |
| TR4 | 101°12′26″ | 41°58′45″ | 924 | Wetland |
| TR5 | 101°16′25″ | 42°01′55″ | 921 | Wetland |
| Sample | CR (No.) | Q30 (%) | GC (%) | Ug (No.) | ML (bp) | N50 (bp) |
|---|---|---|---|---|---|---|
| TR1 | 47893646 | 93.12 | 41.95 | 37790 | 1053.09 | 1953 |
| TR2 | 43861774 | 93.06 | 41.96 | 35818 | 1083.87 | 1983 |
| TR3 | 46194046 | 93.04 | 42.33 | 39252 | 995.67 | 1812 |
| TR4 | 44817976 | 93.17 | 42.87 | 32175 | 1099.48 | 1899 |
| TR5 | 44190698 | 92.92 | 42.49 | 34364 | 1082.91 | 1930 |
| All-unigene | 81728 | 823.46 | 1364 |
Table 2 Statistics of sequencing and assembly results
| Sample | CR (No.) | Q30 (%) | GC (%) | Ug (No.) | ML (bp) | N50 (bp) |
|---|---|---|---|---|---|---|
| TR1 | 47893646 | 93.12 | 41.95 | 37790 | 1053.09 | 1953 |
| TR2 | 43861774 | 93.06 | 41.96 | 35818 | 1083.87 | 1983 |
| TR3 | 46194046 | 93.04 | 42.33 | 39252 | 995.67 | 1812 |
| TR4 | 44817976 | 93.17 | 42.87 | 32175 | 1099.48 | 1899 |
| TR5 | 44190698 | 92.92 | 42.49 | 34364 | 1082.91 | 1930 |
| All-unigene | 81728 | 823.46 | 1364 |
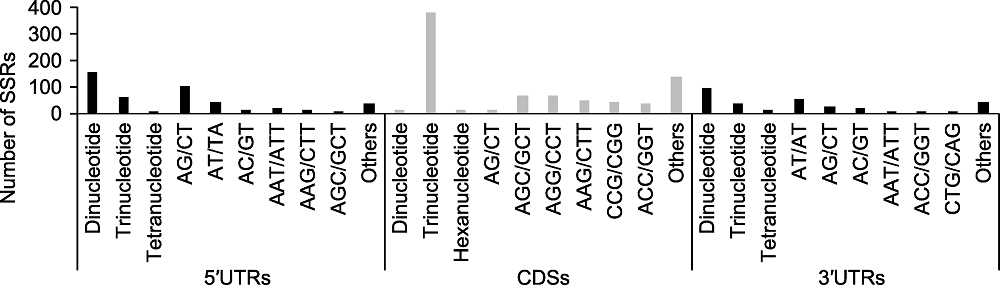
Figure 1 Distribution of the identified Genic-SSRs The x-axis represents the distribution and motif types of polymorphic Genic-SSRs; The y-axis represents the number of Genic- SSRs.
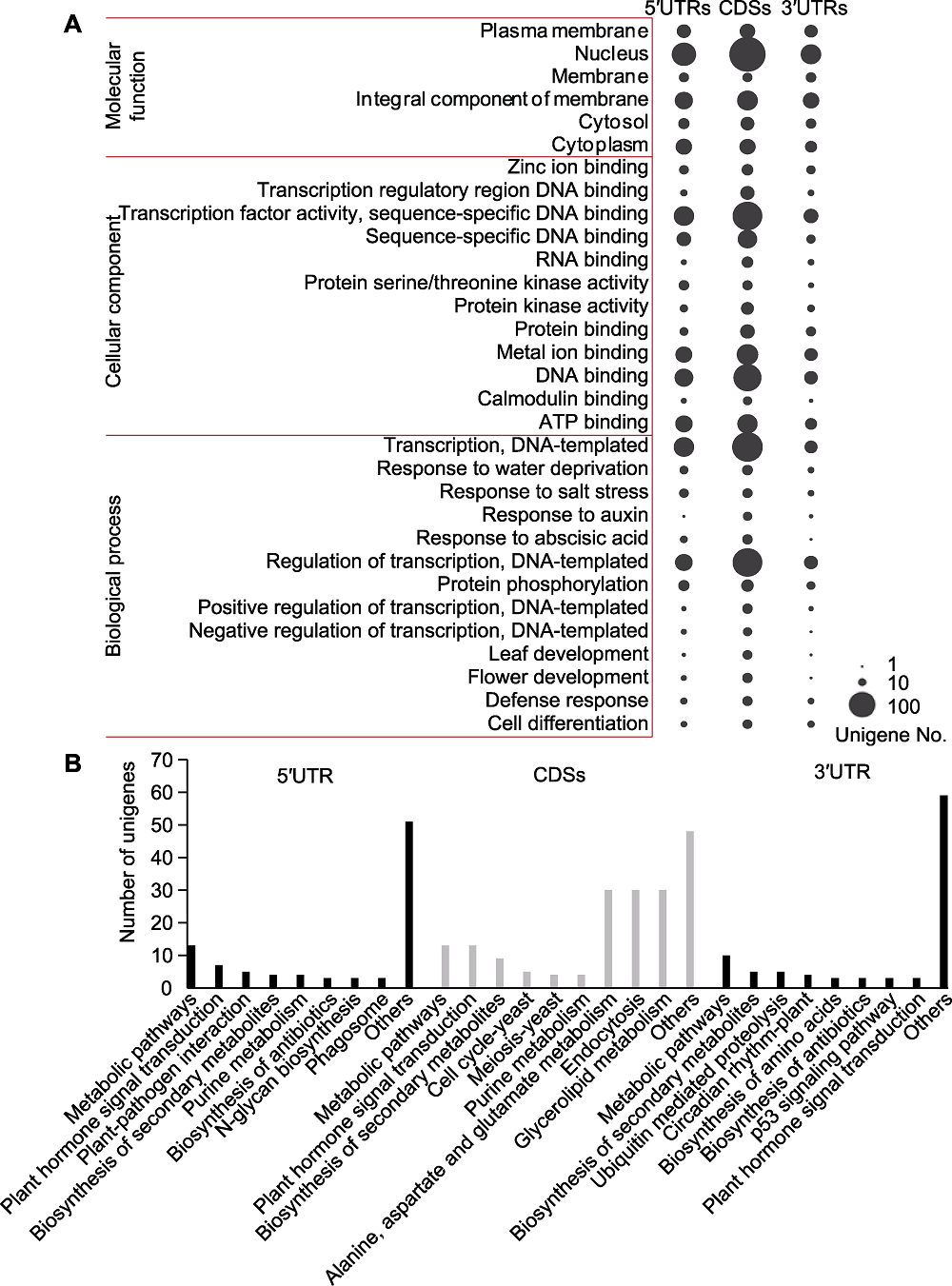
Figure 2 GO and KEGG enrichment of the Genic-SSR-containing sequences (A) GO analysis, the three columns of the bubbles represent GO enrichment analysis of transcripts that containing Genic-SSRs in the 5′UTRs, CDSs and 3′UTRs, respectively, and the number of unigenes assigned to each term is indicated by the size of each bubble; GO terms that contained unigenes more than or equal to ten in one of the gene regions are shown; (B) KEGG enrichment analysis, the x-axis indicates the enriched pathways assigned to the 5°UTRs, CDSs and 3°UTRs Genic-SSR-containing sequences; the y-axis represents the number of unigenes enriched in KEGG pathways.
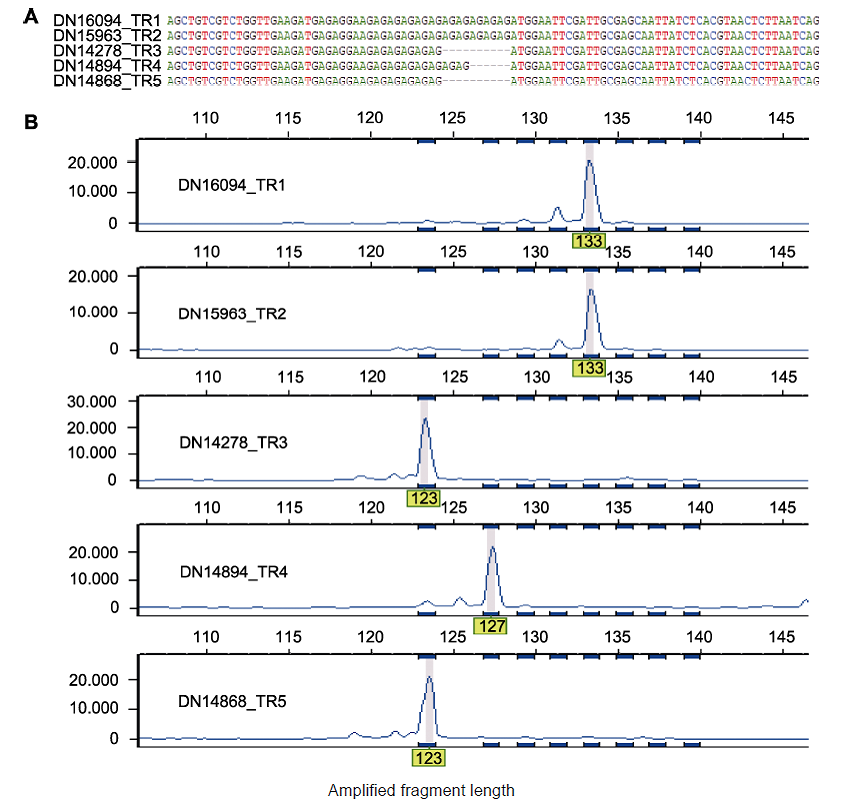
Figure 3 Multiple sequence alignment of polymorphic Genic-SSR and capillary electrophoresis analysis (A) Multiple sequence alignment for five transcripts assembled in the TR1-TR5 transcriptomes that correspond to polymorphic Genic-SSR 104; (B) The polymorphism of Genic-SSRs in (A) revealed by capillary electrophoresis
| SSR ID | PS (5′→3′) | RM | Tm (°C) | AS (bp) | Na | PIC | Ho | He | |
|---|---|---|---|---|---|---|---|---|---|
| 104 | F: CAAGGAGGAGCTGTCGTCTG | AG | 59.9 | 123-139 | 8 | 0.516 | 0.286 | 0.558 | |
| R: GCAGACACGAAGTTTGCGAT | |||||||||
| 221 | F: TGAAGCAGCTGTGTTGGTGA | AT | 53.4 | 158-162 | 3 | 0.562 | 0.321 | 0.645 | |
| R: TCCTCCTCGATTCCCTACTGA | |||||||||
| 300 | F: GAAGGGTTTGGGTGTTTTCAGA | AT | 59.5 | 155-161 | 4 | 0.641 | 0.643 | 0.708 | |
| R: AAAACGCACCCTCTCAGCAG | |||||||||
| 392 | F: CGCAACAAGCACAACATCCA | CAG | 57.9 | 100-109 | 4 | 0.407 | 0.608 | 0.489 | |
| R: GTTAACCGGTCGCACAACTG | |||||||||
| 438 | F: GGCACCGATACACAAGGACA | CCTT | 55.4 | 192-204 | 4 | 0.512 | 0.500 | 0.593 | |
| R: CCCACAGGCTTACACCCATT | |||||||||
| 441 | F: GCCACCGCCACATTTATCTT | CGAG | 50.8 | 106-122 | 5 | 0.520 | 0.286 | 0.570 | |
| R: GGAGCTTGACAGGTACAGCA | |||||||||
| 563 | F: TCCTGTGCAACGAACTGAGT | GA | 51.7 | 176-186 | 6 | 0.593 | 0.536 | 0.666 | |
| R: GGGTTTAGCCATGGGTGACA | |||||||||
| 668 | F: ATGGCGATGATGGAGCAACA | GAG | 59.9 | 146-173 | 7 | 0.711 | 0.429 | 0.758 | |
| R: AGGGATTGGCGGAAGTGAAG | |||||||||
| 670 | F: GAAGACACAGCACCAAGGGA | GAG | 56.7 | 186-201 | 5 | 0.546 | 0.714 | 0.627 | |
| R: CGTCTTCCCCAAGTCCGATC | |||||||||
| 947 | F: TCCCCACACGTAATCCCTTC | TC | 50.8 | 139-149 | 5 | 0.445 | 0.607 | 0.488 | |
| R: GCGGATGGAAGGAGAAGAGG | |||||||||
| 1024 | F: TGCGCATTTCTTGATTGCCA | TCT | 52.1 | 168-180 | 5 | 0.476 | 0.464 | 0.516 | |
| R: GGGTTGATGCGGCTTGATTG | |||||||||
| 1053 | F: GTCGACACTGCAAGCATCAC | TG | 56.7 | 202-204 | 2 | 0.293 | 0.036 | 0.363 | |
| R: GCACGTACGAGCACATCTCT | |||||||||
| 1093 | F: GAATAGTGGTGGCGGGAGTC | TGC | 54.7 | 117-126 | 2 | 0.195 | 0.250 | 0.223 | |
| R: GCTTGGCATCGTACCCCTAT | |||||||||
| 1114 | F: GGTCCTCAGTGTGGCCATAG | TGG | 54.1 | 156-171 | 4 | 0.487 | 0.214 | 0.543 | |
| R: GTGTTAGAGATGGCGGGCAA | |||||||||
| Mean | 4.5 | 0.493 | 0.421 | 0.553 | |||||
Table 3 Basic information and genetic parameters of the 14 polymorphic Genic-SSRs
| SSR ID | PS (5′→3′) | RM | Tm (°C) | AS (bp) | Na | PIC | Ho | He | |
|---|---|---|---|---|---|---|---|---|---|
| 104 | F: CAAGGAGGAGCTGTCGTCTG | AG | 59.9 | 123-139 | 8 | 0.516 | 0.286 | 0.558 | |
| R: GCAGACACGAAGTTTGCGAT | |||||||||
| 221 | F: TGAAGCAGCTGTGTTGGTGA | AT | 53.4 | 158-162 | 3 | 0.562 | 0.321 | 0.645 | |
| R: TCCTCCTCGATTCCCTACTGA | |||||||||
| 300 | F: GAAGGGTTTGGGTGTTTTCAGA | AT | 59.5 | 155-161 | 4 | 0.641 | 0.643 | 0.708 | |
| R: AAAACGCACCCTCTCAGCAG | |||||||||
| 392 | F: CGCAACAAGCACAACATCCA | CAG | 57.9 | 100-109 | 4 | 0.407 | 0.608 | 0.489 | |
| R: GTTAACCGGTCGCACAACTG | |||||||||
| 438 | F: GGCACCGATACACAAGGACA | CCTT | 55.4 | 192-204 | 4 | 0.512 | 0.500 | 0.593 | |
| R: CCCACAGGCTTACACCCATT | |||||||||
| 441 | F: GCCACCGCCACATTTATCTT | CGAG | 50.8 | 106-122 | 5 | 0.520 | 0.286 | 0.570 | |
| R: GGAGCTTGACAGGTACAGCA | |||||||||
| 563 | F: TCCTGTGCAACGAACTGAGT | GA | 51.7 | 176-186 | 6 | 0.593 | 0.536 | 0.666 | |
| R: GGGTTTAGCCATGGGTGACA | |||||||||
| 668 | F: ATGGCGATGATGGAGCAACA | GAG | 59.9 | 146-173 | 7 | 0.711 | 0.429 | 0.758 | |
| R: AGGGATTGGCGGAAGTGAAG | |||||||||
| 670 | F: GAAGACACAGCACCAAGGGA | GAG | 56.7 | 186-201 | 5 | 0.546 | 0.714 | 0.627 | |
| R: CGTCTTCCCCAAGTCCGATC | |||||||||
| 947 | F: TCCCCACACGTAATCCCTTC | TC | 50.8 | 139-149 | 5 | 0.445 | 0.607 | 0.488 | |
| R: GCGGATGGAAGGAGAAGAGG | |||||||||
| 1024 | F: TGCGCATTTCTTGATTGCCA | TCT | 52.1 | 168-180 | 5 | 0.476 | 0.464 | 0.516 | |
| R: GGGTTGATGCGGCTTGATTG | |||||||||
| 1053 | F: GTCGACACTGCAAGCATCAC | TG | 56.7 | 202-204 | 2 | 0.293 | 0.036 | 0.363 | |
| R: GCACGTACGAGCACATCTCT | |||||||||
| 1093 | F: GAATAGTGGTGGCGGGAGTC | TGC | 54.7 | 117-126 | 2 | 0.195 | 0.250 | 0.223 | |
| R: GCTTGGCATCGTACCCCTAT | |||||||||
| 1114 | F: GGTCCTCAGTGTGGCCATAG | TGG | 54.1 | 156-171 | 4 | 0.487 | 0.214 | 0.543 | |
| R: GTGTTAGAGATGGCGGGCAA | |||||||||
| Mean | 4.5 | 0.493 | 0.421 | 0.553 | |||||
| [1] | 毕江涛, 马萍, 杨志伟, 关晓庆 (2013). 药用植物柽柳内生真菌分离及其抑菌活性初步研究. 草业学报 22(3), 132-138. |
| [2] |
陈敏, 刘林德, 张莉, 王丽娟 (2012). 黑河中游和烟台海滨中国柽柳的传粉生态学研究. 植物学报 47, 264-270.
DOI |
| [3] | 陈雨, 潘大建, 曲延英, 范芝兰, 陈建酉, 李晨 (2008). 广东高州7个普通野生稻居群遗传结构的SSR分析. 植物学通报 25, 430-436. |
| [4] | 蒋志敏, 陈玉霞, 包颖 (2011). 黄河三角洲柽柳居群的遗传结构和遗传分化. 植物分类与资源学报 33, 403-408. |
| [5] | 李珊, 周天华, 赵桂仿, 朱云国, 杨晓伶, 程舟 (2010). 马蹄香表达序列标签资源的SSR信息分析. 中草药 41, 464-468. |
| [6] | 李永涛, 王霞, 魏海霞, 杨庆山, 周健, 刘德玺, 刘忠杰, 魏文千 (2017). 盐碱生境模拟下两种柽柳的生理特性研究. 山东农业科学 49, 53-58. |
| [7] | 刘林, 李进斌, 张悦, 苏源, 杨静, 陈梦琪, 李成云 (2010). 灰色大角间座壳菌(稻瘟病菌)蛋白激酶编码基因中SSR变异及其对蛋白结构的潜在影响. 菌物学报 29, 698-706. |
| [8] | 王慧, 刘宁, 刘金龙, 姚延梼, 王林 (2020). 晋北干旱区盐碱地柽柳形态特征及其与土壤养分的关系. 中南林业科技大学学报 40, 37-48. |
| [9] | 温月仙, 甘红豪, 史胜青, 江泽平, 吴利禄, 褚建民 (2020). 基于叶绿体和核基因片段序列的甘蒙柽柳谱系地理研究. 林业科学 56, 55-64. |
| [10] | 伍明江, 张德芹, 李盼, 石旭柳, 刘璐 (2020). 柽柳黄素对3T3-L1脂肪细胞胰岛素抵抗的影响及AMPK信号通路的作用机制. 天然产物研究与开发 32, 953-960. |
| [11] | 杨成君, 王军 (2008). 人参EST资源的SSR信息分析. 植物生理学通讯 44, 69-73. |
| [12] | 姚秋阳 (2015). 利用RNA-seq技术在云南山茶中解析重要分子通路与开发多态性EST-SSR. 博士论文. 昆明: 云南大学. pp. 38-96. |
| [13] | 叶春秀, 姜继元, 董鹏, 庄振刚, 陈奇凌, 谢宗铭 (2015). 基于SSR标记的新疆塔里木河流域柽柳指纹图谱构建及遗传多样性分析. 分子植物育种 13, 2566-2571. |
| [14] |
Bräutigam A, Gowik U (2010). What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol 12, 831-841.
DOI URL |
| [15] |
Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R (2000). Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156, 847-854.
PMID |
| [16] |
Chen CX, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006). Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112, 1248-1257.
DOI URL |
| [17] |
Chen YN, Zhou HH, Chen YP (2013). Adaptation strategies of desert riparian forest vegetation in response to drought stress. Ecohydrology 6, 956-973.
DOI URL |
| [18] |
Dang ZH, Huang L, Jia YY, Lockhart PJ, Fong Y, Tian YY (2020). Identification of Genic SSRs provide a perspective for studying environmental adaptation in the endemic shrub Tetraena mongolica. Genes 11, 322.
DOI URL |
| [19] |
Fraser LG, Harvey CF, Crowhurst RN, Silva HN (2004). EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108, 1010-1016.
PMID |
| [20] |
Gao LF, Tang JF, Li HW, Jia JZ (2003). Analysis of microsatellites in major crops assessed by computational and experimental approaches. Mol Breed 12, 245-261.
DOI URL |
| [21] |
Gou XJ, Shi HR, Yu SF, Wang ZQ, Li CX, Liu SH, Ma J, Chen GD, Liu T, Liu YX (2020). SSRMMD: a rapid and accurate algorithm for mining SSR feature loci and candidate polymorphic SSRs based on assembled sequences. Front Genet 11, 706.
DOI URL |
| [22] |
Han ZZ, Ma XY, Wei M, Zhao T, Zhan RT, Chen WW (2018). SSR marker development and intraspecific genetic divergence exploration of Chrysanthemum indicum based on transcriptome analysis. BMC Genomics 19, 291.
DOI URL |
| [23] |
Li YC, Korol AB, Fahima T, Beiles A, Nevo E (2002). Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11, 2453-2465.
DOI URL |
| [24] |
Li YC, Korol AB, Fahima T, Nevo E (2004). Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21, 991-1007.
DOI URL |
| [25] |
Li ZT, Zhong YD, Yu FX, Xu M (2018). Novel SSR marker development and genetic diversity analysis of Cinnamomum camphora based on transcriptome sequencing. Plant Genet Res 16, 568-571.
DOI URL |
| [26] |
Liu KJ, Muse SV (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128-2129.
DOI URL |
| [27] |
Liu LY, Fan XF, Tan PH, Wu JY, Zhang H, Han C, Chen C, Xun LL, Guo WE, Chang ZH, Teng K (2021). The development of SSR markers based on RNA-sequencing and its validation between and within Carex L. species. BMC Plant Biol 21, 17.
DOI URL |
| [28] | Lynch M, Lande R (1993). Biotic Interactions and Global Change. Sunderland MA: Sinauer Assocs.Inc. pp. 234-250. |
| [29] |
Morgante M, Hanafey M, Powell W (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30, 194-200.
PMID |
| [30] |
Natale E, Zalba SM, Oggero A, Reinoso H (2010). Establishment of Tamarix ramosissima under different conditions of salinity and water availability: implications for its management as an invasive species. J Arid Environ 74, 1399-1407.
DOI URL |
| [31] |
Qin LP, Wang LQ, Guo Y, Li Y, Ümüt H, Wang YC (2017). An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance. Plant Sci 265, 154-166.
DOI URL |
| [32] |
Ranathunge C, Wheeler GL, Chimahusky ME, Perkins AD, Pramod S, Welch ME (2020). Transcribed microsatellite allele lengths are often correlated with gene expression in natural sunflower populations. Mol Ecol 29, 1704-1716.
DOI URL |
| [33] |
Santi L, Wang YM, Stile MR, Berendzen K, Wanke D, Roig C, Pozzi C, Müller K, Müller J, Rohde W, Salamini F (2003). The GA octodinucleotide repeat binding factor BBR participates in the transcriptional regulation of the homeobox gene Bkn3. Plant J 34, 813-826.
PMID |
| [34] |
Sawaya S, Bagshaw A, Buschiazzo E, Kumar P, Chowdhury S, Black MA, Gemmell N (2013). Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS One 8, e54710.
DOI URL |
| [35] |
Trifonov EN (1995). Segmented structure of protein sequences and early evolution of genome by combinatorial fusion of DNA elements. J Mol Evol 40, 337-342.
PMID |
| [1] | Yaping Wang, Wenquan Bao, Yu’e Bai. Advances in the Application of Single-cell Transcriptomics in Plant Growth, Development and Stress Response [J]. Chinese Bulletin of Botany, 2025, 60(1): 101-113. |
| [2] | Chunjiao Xia, Yunguang Li, Shu Xia, Wei Pang, Chunli Chen. Flow Cytometric Analysis and Sorting in Plant Genomics [J]. Chinese Bulletin of Botany, 2024, 59(5): 774-782. |
| [3] | CHENG Ke-Xin, DU Yao, LI Kai-Hang, WANG Hao-Chen, YANG Yan, JIN Yi, HE Xiao-Qing. Genetic mechanism of interaction between maize and phyllospheric microbiome [J]. Chin J Plant Ecol, 2024, 48(2): 215-228. |
| [4] | Zumureti YUSUFUJANG, DONG Zheng-Wu, CHENG Peng, YE Mao, LIU Sui-Yun-Hao, LI Sheng-Yu, ZHAO Xiao-Ying. Response of water use strategies of Tamarix ramosissima to nebkhas accumulation process [J]. Chin J Plant Ecol, 2024, 48(1): 113-126. |
| [5] | Wei Heping, Lu Tao, Jia Qiwei, Deng Fei, Zhu Hao, Qi Zehua, Wang Yuxi, Ye Hanfei, Yin Wenjing, Fang Yuan, Mu Dan, Rao Yuchun. QTL Mapping of Candidate Genes for Heading Date in Rice [J]. Chinese Bulletin of Botany, 2022, 57(5): 588-595. |
| [6] | Haixia Xu, Jing He, Hang Yi, Li Wang. Sex Specific Response Mechanism of Transcriptome in Both Male and Female Marchantia polymorpha Under Cadmium Stress [J]. Chinese Bulletin of Botany, 2022, 57(2): 182-196. |
| [7] | Gang Ren, En Li, Shiye Zhao, Yanqiong Jiang, Shasha Wang, Sixian Tang, Huijian Hu. Correlation between color polymorphism and the MC1R gene of Lanius schach [J]. Biodiv Sci, 2020, 28(6): 688-694. |
| [8] | Xi Zhang, Tianhang Qiu, Anan Wang, Huajian Zhou, Min Yuan, Li Li, Sulan Bai, Suxia Cui. Morphology and Genetic Diversity of Phragmites australis in Beijing [J]. Chinese Bulletin of Botany, 2020, 55(6): 693-704. |
| [9] | Yan Yang, Haiqin Zhang, Xing Fan, Lina Sha, Houyang Kang, Yi Wang, Yonghong Zhou. Polymorphism of Gliadin and Glutelin and Systematics Studies in Elytrigia [J]. Chinese Bulletin of Botany, 2017, 52(5): 579-589. |
| [10] | Jianfeng Huang, Lang Li, Jie Li. Polymorphism of the Internal Transcribed Spacer of nrDNA in Cinnamomum Schaeffer (Lauraceae) [J]. Chinese Bulletin of Botany, 2016, 51(5): 609-619. |
| [11] | Yanyan Lü, Sanxiong Fu, Song Chen, Wei Zhang, Cunkou Qi. RNA-sequencing Analysis of Differentially Expressed Genes in Wild-type and BnERF-transgenic Arabidopsis Under Submergence Treatment [J]. Chinese Bulletin of Botany, 2015, 50(3): 321-330. |
| [12] | Wen Fan, Ying Xu, Ting Xu, Jing Xu, Takahiro Yonezawa, Jiyin Gao, Wenju Zhang. Intragenomic Polymorphism of the Internal Transcribed Spacer Region of Ribosomal DNA in Camellia hongkongensis (Theaceae) and Species Identification [J]. Chinese Bulletin of Botany, 2015, 50(2): 217-226. |
| [13] | WANG Na-Na, CHEN Ying, YING Jiao-Yan, GAO Yong-Sheng, BAI Yong-Fei. Effects of typical plant on soil microbial communities in an Inner Mongolia grassland [J]. Chin J Plant Ecol, 2014, 38(2): 201-208. |
| [14] | Yuanfeng Cai,Zhongjun Jia. Progress in environmental transcriptomics based on next-generation high-throughput sequencing [J]. Biodiv Sci, 2013, 21(4): 401-410. |
| [15] | MA Xiao-Dong, ZHU Cheng-Gang, LI Wei-Hong. Response of root morphology and biomass of Tamarix ramosissima seedlings to different water irrigations [J]. Chin J Plant Ecol, 2012, 36(10): 1024-1032. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||