

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (6): 733-743.DOI: 10.11983/CBB18235 cstr: 32102.14.CBB18235
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Na Zhang1,2,Xiuxia Liu1,Xuesen Chen1,*( ),Shujing Wu1,*(
),Shujing Wu1,*( )
)
Received:2018-11-04
Accepted:2019-02-11
Online:2019-11-01
Published:2020-07-09
Contact:
Xuesen Chen,Shujing Wu
Na Zhang,Xiuxia Liu,Xuesen Chen,Shujing Wu. Identifying Genes Responsive to Jasmonates in Apple Based on Transcriptome Analysis[J]. Chinese Bulletin of Botany, 2019, 54(6): 733-743.
| Sample | Total reads | Total base pairs | Total mapped reads | Perfect match | Mismatch | Unique match | Muti-position match | Total unmapped reads |
|---|---|---|---|---|---|---|---|---|
| CTRL1 | 15336268 (100.00%) | 751477132 (100.00%) | 10932142 (71.28%) | 6783455 (44.23%) | 4148687 (27.05%) | 9149425 (59.66%) | 1782717 (11.62%) | 4404126 (28.72%) |
| MeJA | 15339040 (100.00%) | 751612960 (100.00%) | 11052987 (72.06%) | 6921067 (45.12%) | 4131920 (26.94%) | 9177064 (59.83%) | 1875923 (12.23%) | 4286053 (27.94%) |
Table 1 The alignment result of genome sequences between sample (Gala) and Golden Delicious apple
| Sample | Total reads | Total base pairs | Total mapped reads | Perfect match | Mismatch | Unique match | Muti-position match | Total unmapped reads |
|---|---|---|---|---|---|---|---|---|
| CTRL1 | 15336268 (100.00%) | 751477132 (100.00%) | 10932142 (71.28%) | 6783455 (44.23%) | 4148687 (27.05%) | 9149425 (59.66%) | 1782717 (11.62%) | 4404126 (28.72%) |
| MeJA | 15339040 (100.00%) | 751612960 (100.00%) | 11052987 (72.06%) | 6921067 (45.12%) | 4131920 (26.94%) | 9177064 (59.83%) | 1875923 (12.23%) | 4286053 (27.94%) |
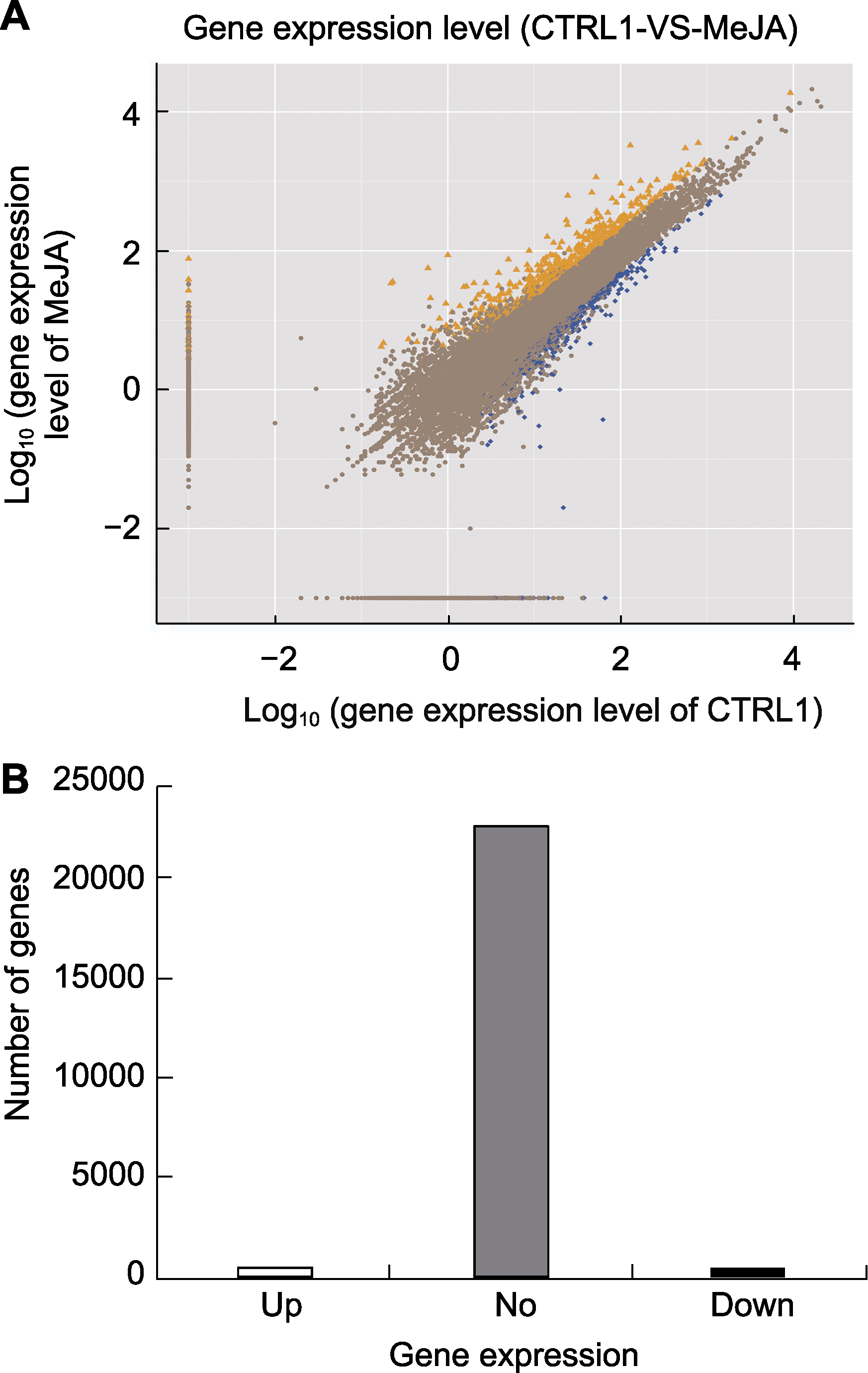
Figure 1 The screening of differentially expressed genes in leaves of apple treated by MeJA (A) Gene expression level. X-axis represents log10 (gene expression level of CTRL1), Y-axis represents log10 (gene expression level of MeJA) (The orange triangle represents up-regulated genes, the blue block represents down-regu- lated genes, and the brown dot represents no significant difference genes); (B) Differentially expressed genes. Up: Up-regulated genes; No: No significant difference genes; Down: Down-regulated genes
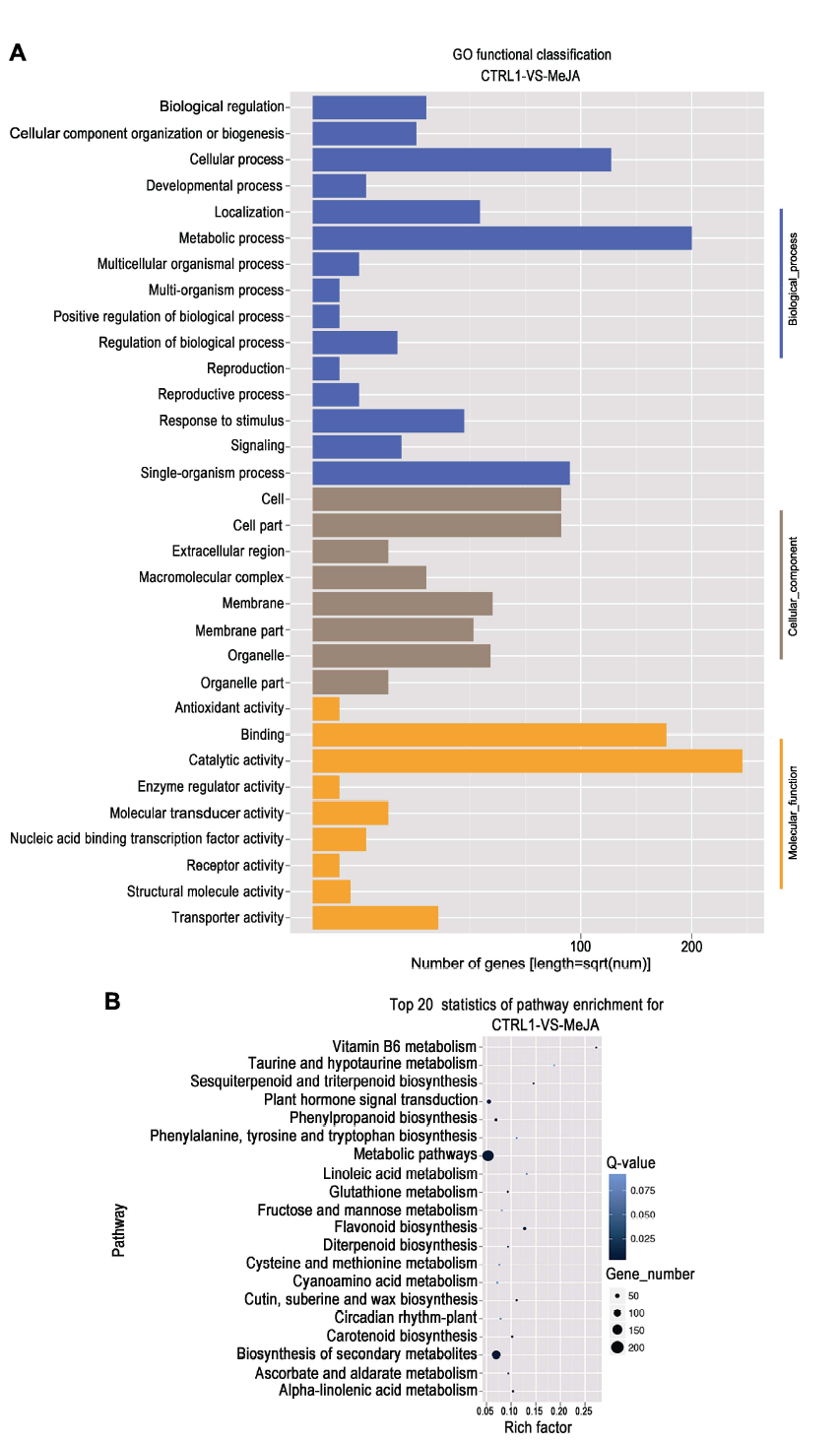
Figure 2 GO functional classification (A) and KEGG enrichment classification (B) of differentially expressed genes in apple leaves under MeJA treatment
| Gene ID | Log2 ratio (MeJA/CTRL1) | KEGG orthology | Blast nr | Pathway |
|---|---|---|---|---|
| MDP0000702120 | 12.32 | K14181 | (-)-alpha-pinene synthase-like | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000205617 | 7.28 | K14181 | Putative pinene synthase | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000919962 | 6.57 | K14181 | (-)-alpha-pinene synthase-like | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000225361 | 3.72 | K14181 | (-)-germacrene D synthase-like | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000248152 | 3.24 | K14181 | (-)-germacrene D synthase-like | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000692178 | 12.12 | K00517 | Cytochrome P450 82G1-like | Flavonoid biosynthesis |
| MDP0000503940 | 4.67 | K05277 | Protein SRG1-like | Flavonoid biosynthesis |
| MDP0000225939 | 3.55 | K05277 | Flavonol synthase/flavanone 3-hydroxylase-like | Flavonoid biosynthesis |
| MDP0000523477 | -3.17 | K01859 | Chalcone-flavonone isomerase-like isoform X5 | Flavonoid biosynthesis |
| MDP0000317152 | 13.53 | K13407 | Cytochrome P450 94C1-like | Cutin, suberine and wax biosynthesis |
| MDP0000153822 | 3.25 | K13407 | Cytochrome P450 94B3-like | Cutin, suberine and wax biosynthesis |
| MDP0000218691 | 3.89 | K01183 | Acidic endochitinase-like | Amino sugar and nucleotide sugar metabolism |
| MDP0000280265 | 3.22 | K01183 | Acidic endochitinase-like | Amino sugar and nucleotide sugar metabolism |
| MDP0000687619 | 12.96 | K09755 | Cytochrome P450 CYP736A12-like | Phenylpropanoid biosynthesis |
| MDP0000539956 | 3.9 | K09755 | Cytochrome P450 CYP736A12-like | Phenylpropanoid biosynthesis |
| MDP0000313394 | 6.44 | K13267 | Cytochrome P450 CYP736A12-like | Isoflavonoid biosynthesis |
| MDP0000813805 | -6.29 | K09840 | 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like | Carotenoid biosynthesis |
| MDP0000228070 | -5.24 | K09840 | 9-cis-epoxycarotenoid dioxygenase NCED1, chloroplastic-like | Carotenoid biosynthesis |
| Gene ID | Log2 ratio (MeJA/CTRL1) | KEGG orthology | Blast nr | Pathway |
|---|---|---|---|---|
| MDP0000702120 | 12.32 | K14181 | (-)-alpha-pinene synthase-like | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000205617 | 7.28 | K14181 | Putative pinene synthase | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000919962 | 6.57 | K14181 | (-)-alpha-pinene synthase-like | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000225361 | 3.72 | K14181 | (-)-germacrene D synthase-like | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000248152 | 3.24 | K14181 | (-)-germacrene D synthase-like | Sesquiterpenoid and triterpenoid biosynthesis |
| MDP0000692178 | 12.12 | K00517 | Cytochrome P450 82G1-like | Flavonoid biosynthesis |
| MDP0000503940 | 4.67 | K05277 | Protein SRG1-like | Flavonoid biosynthesis |
| MDP0000225939 | 3.55 | K05277 | Flavonol synthase/flavanone 3-hydroxylase-like | Flavonoid biosynthesis |
| MDP0000523477 | -3.17 | K01859 | Chalcone-flavonone isomerase-like isoform X5 | Flavonoid biosynthesis |
| MDP0000317152 | 13.53 | K13407 | Cytochrome P450 94C1-like | Cutin, suberine and wax biosynthesis |
| MDP0000153822 | 3.25 | K13407 | Cytochrome P450 94B3-like | Cutin, suberine and wax biosynthesis |
| MDP0000218691 | 3.89 | K01183 | Acidic endochitinase-like | Amino sugar and nucleotide sugar metabolism |
| MDP0000280265 | 3.22 | K01183 | Acidic endochitinase-like | Amino sugar and nucleotide sugar metabolism |
| MDP0000687619 | 12.96 | K09755 | Cytochrome P450 CYP736A12-like | Phenylpropanoid biosynthesis |
| MDP0000539956 | 3.9 | K09755 | Cytochrome P450 CYP736A12-like | Phenylpropanoid biosynthesis |
| MDP0000313394 | 6.44 | K13267 | Cytochrome P450 CYP736A12-like | Isoflavonoid biosynthesis |
| MDP0000813805 | -6.29 | K09840 | 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like | Carotenoid biosynthesis |
| MDP0000228070 | -5.24 | K09840 | 9-cis-epoxycarotenoid dioxygenase NCED1, chloroplastic-like | Carotenoid biosynthesis |
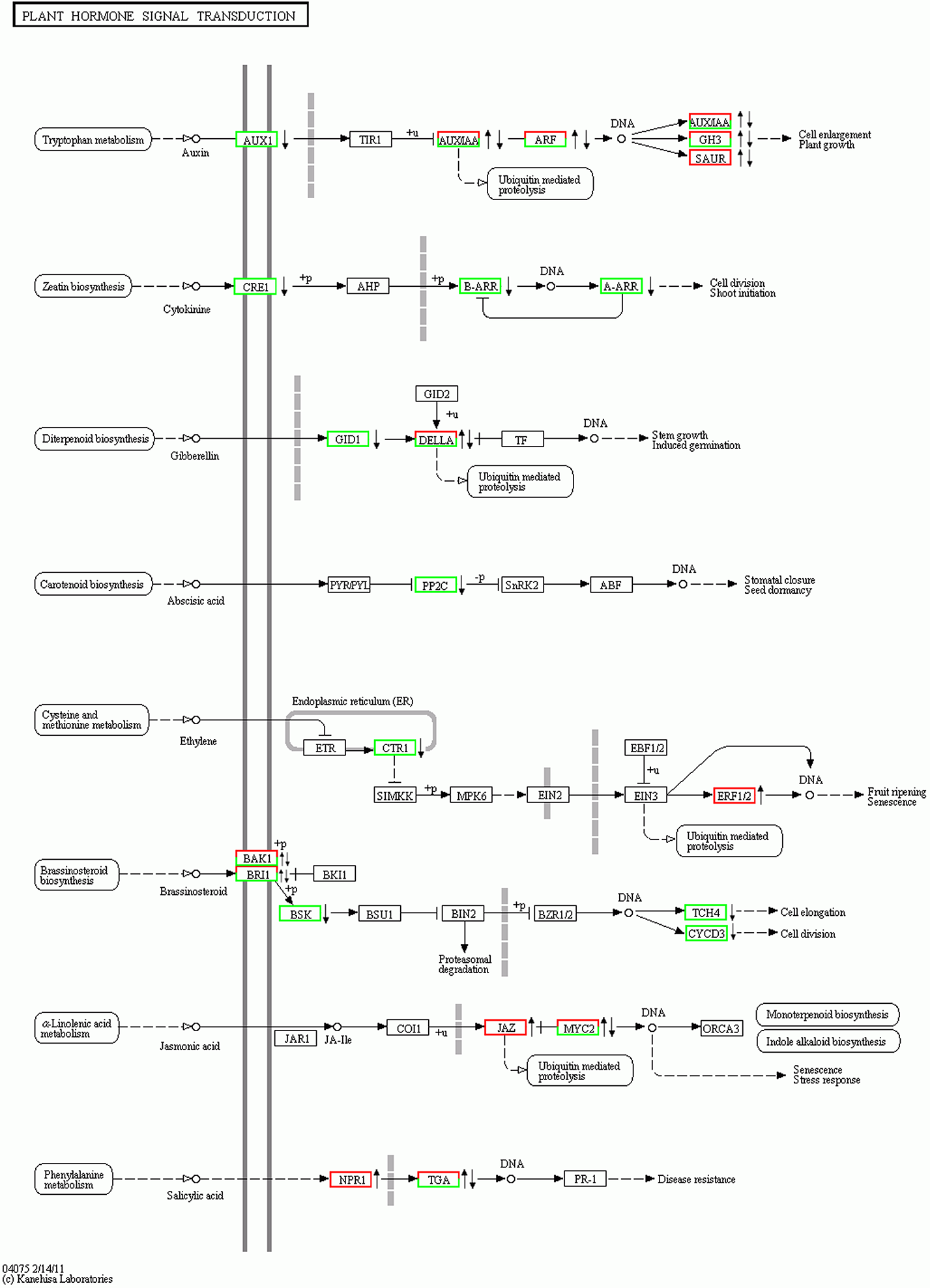
Figure 3 Effect of MeJA on plant hormone signaling pathways in leaves of apple on the right side of red frame: Up-regulated; on the right side of green frame: Down-regulated; on the right side of red-green frame: Include both up-regulated and down-regulated genes.
| Plant hormone signaling pathway | Gene ID | Log2 ratio (MeJA/CTRL1) |
|---|---|---|
| Auxin | MDP0000300452 | -1.4 |
| MDP0000255223 MDP0000213864 | 1.1 -1.1 | |
| MDP0000143749 MDP0000221322 | 1.3 -1.6 | |
| MDP0000873893 MDP0000214081 | 1.5 -1.1 | |
| MDP0000786165 | 1.1 | |
| Cytokinine | MDP0000146863 | -1.1 |
| MDP0000285242 MDP0000225179 | 1.6 -1.0 | |
| MDP0000250737 | -1.5 | |
| Gibberellin | MDP0000227827 MDP0000868088 | -1.3 -1.0 |
| MDP0000284679 MDP0000227056 MDP0000397638 | 1.4 1.3 -2.2 | |
| Abscisic acid | MDP0000437033 MDP0000203818 MDP0000283274 | -1.6 -1.3 -1.2 |
| Ethylene | MDP0000275915 | -2.4 |
| MDP0000855671 MDP0000127134 MDP0000235313 MDP0000805422 MDP0000787281 | 3.1 2.5 1.8 1.2 1.0 | |
| Brassinosteroid | MDP0000123379 MDP0000319460 MDP0000319708 MDP0000904826 MDP0000196862 | 12.5 1.9 1.0 1.0 -1.8 |
| MDP0000217213 MDP0000897962 MDP0000290950 MDP0000620422 MDP0000303744 MDP0000189315 MDP0000741253 MDP0000223726 MDP0000189841 | 1.9 1.3 -2.3 -1.8 -1.6 -1.1 -1.0 -1.0 -1.0 | |
| MDP0000130143 | -12.8 | |
| MDP0000320017 | -1.4 | |
| MDP0000135392 | -1.0 | |
| Jasmonic acid | MDP0000565690 MDP0000187921 MDP0000889413 MDP0000301927 | 2.8 2.6 1.7 1.2 |
| MDP0000603546 MDP0000242554 MDP0000029168 MDP0000900024 MDP0000226497 MDP0000290263 | 3.6 2.2 1.6 1.6 1.0 -1.0 | |
| Salicylic acid | MDP0000292425 | 1.8 |
| MDP0000529682 MDP0000375992 | 1.1 -4.3 |
Table 3 The significant differentially expressed genes involved in plant hormone signaling in leaves of apple under MeJA treatment
| Plant hormone signaling pathway | Gene ID | Log2 ratio (MeJA/CTRL1) |
|---|---|---|
| Auxin | MDP0000300452 | -1.4 |
| MDP0000255223 MDP0000213864 | 1.1 -1.1 | |
| MDP0000143749 MDP0000221322 | 1.3 -1.6 | |
| MDP0000873893 MDP0000214081 | 1.5 -1.1 | |
| MDP0000786165 | 1.1 | |
| Cytokinine | MDP0000146863 | -1.1 |
| MDP0000285242 MDP0000225179 | 1.6 -1.0 | |
| MDP0000250737 | -1.5 | |
| Gibberellin | MDP0000227827 MDP0000868088 | -1.3 -1.0 |
| MDP0000284679 MDP0000227056 MDP0000397638 | 1.4 1.3 -2.2 | |
| Abscisic acid | MDP0000437033 MDP0000203818 MDP0000283274 | -1.6 -1.3 -1.2 |
| Ethylene | MDP0000275915 | -2.4 |
| MDP0000855671 MDP0000127134 MDP0000235313 MDP0000805422 MDP0000787281 | 3.1 2.5 1.8 1.2 1.0 | |
| Brassinosteroid | MDP0000123379 MDP0000319460 MDP0000319708 MDP0000904826 MDP0000196862 | 12.5 1.9 1.0 1.0 -1.8 |
| MDP0000217213 MDP0000897962 MDP0000290950 MDP0000620422 MDP0000303744 MDP0000189315 MDP0000741253 MDP0000223726 MDP0000189841 | 1.9 1.3 -2.3 -1.8 -1.6 -1.1 -1.0 -1.0 -1.0 | |
| MDP0000130143 | -12.8 | |
| MDP0000320017 | -1.4 | |
| MDP0000135392 | -1.0 | |
| Jasmonic acid | MDP0000565690 MDP0000187921 MDP0000889413 MDP0000301927 | 2.8 2.6 1.7 1.2 |
| MDP0000603546 MDP0000242554 MDP0000029168 MDP0000900024 MDP0000226497 MDP0000290263 | 3.6 2.2 1.6 1.6 1.0 -1.0 | |
| Salicylic acid | MDP0000292425 | 1.8 |
| MDP0000529682 MDP0000375992 | 1.1 -4.3 |
| Gene ID | Log2 ratio (MeJA/CTRL) | Annotation |
|---|---|---|
| MDP0000123379 | 12.46 | Putative serine/threonine- protein kinase |
| MDP0000603546 | 3.56 | Transcription factor MYC2- like |
| MDP0000855671 | 3.1 | Ethylene-responsive transcription factor 1B-like |
| MDP0000130143 | -12.82 | Probable serine/threonine- protein kinase At5g41260 |
| MDP0000375992 | -4.27 | U-box domain-containing protein 19-like |
Table 4 The regulated genes involved in biosynthesis of secondary metabolites pathway in leaves of apple under MeJA treatment (Log2 (MeJA/CTRL)≥3)
| Gene ID | Log2 ratio (MeJA/CTRL) | Annotation |
|---|---|---|
| MDP0000123379 | 12.46 | Putative serine/threonine- protein kinase |
| MDP0000603546 | 3.56 | Transcription factor MYC2- like |
| MDP0000855671 | 3.1 | Ethylene-responsive transcription factor 1B-like |
| MDP0000130143 | -12.82 | Probable serine/threonine- protein kinase At5g41260 |
| MDP0000375992 | -4.27 | U-box domain-containing protein 19-like |
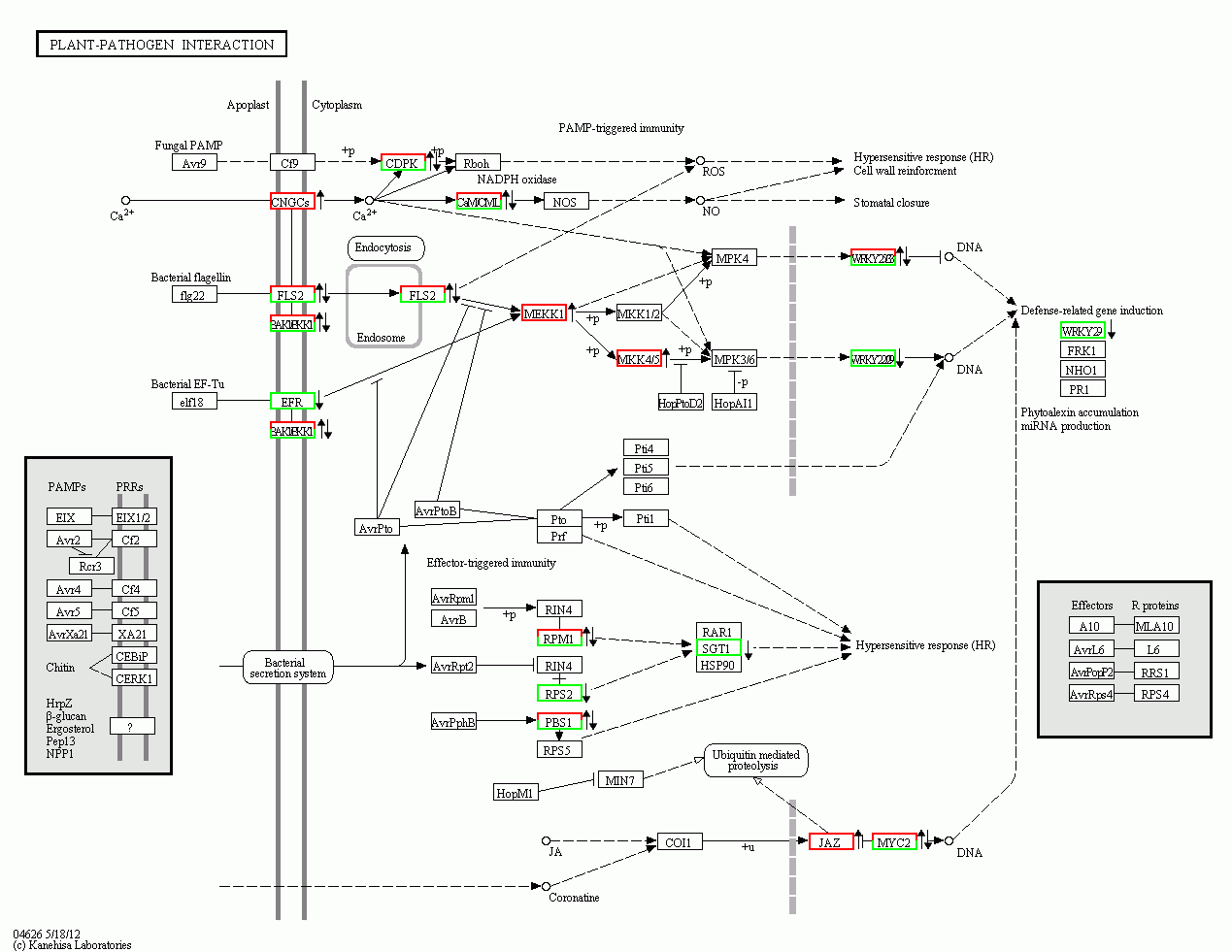
Figure 4 Effect of MeJA on plant-pathogen interaction pathway in leaves of apple on the right side of red frame: Up-regulated; on the right side of green frame: Down-regulated; on the right side of red-green frame: Include both up-regulated and down-regulated genes.
| [1] | 宾金华, 潘瑞炽 ( 1995). 茉莉酸甲酯的生理生化及在植物抗病中的作用. 植物学通报 12(4), 17-21. |
| [2] | 陈学森, 郭文武, 徐娟, 从佩华, 王力荣, 刘崇怀, 李秀根, 吴树敬, 姚玉新, 陈晓流 ( 2015). 主要果树果实品质遗传改良与提升实践. 中国农业科学 48, 3524-3540. |
| [3] | 弓德强, 谷会, 张鲁斌, 洪克前, 朱世江 ( 2013). 杧果采前喷施茉莉酸甲酯对其抗病性和采后品质的影响. 园艺学报 40, 49-57. |
| [4] | 国立耘, 李金云, 李保华, 张新忠, 周增强, 李广旭, 王英姿, 李晓军, 黄丽丽, 孙广宇, 文耀东 ( 2009). 中国苹果枝干轮纹病发生和防治情况. 植物保护 35(4), 120-123. |
| [5] | 李灿婴, 葛永红, 朱丹实, 张惠君 ( 2015). 采后茉莉酸甲酯处理对富士苹果青霉病和贮藏品质的影响. 食品科学 36(2), 255-259. |
| [6] | 李梦莎, 阎秀峰 ( 2014). 植物的环境信号分子茉莉酸及其生物学功能. 生态学报 34, 6779-6788. |
| [7] | 刘志, 张华磊, 谢兴斌, 束怀瑞, 郝玉金 ( 2009). 茉莉酸甲酯诱导的苹果悬浮细胞的防卫响应. 园艺学报 36, 171-178. |
| [8] | 罗昌国, 袁启凤, 裴晓红, 吴亚维, 郑伟, 章镇 ( 2013). 富士苹果MdWRKY40b基因克隆及其对白粉病的抗性分析. 西北植物学报 33, 2382-2387. |
| [9] | 麻宝成, 朱世江 ( 2006). 苯丙噻重氮和茉莉酸甲酯对采后香蕉果实抗病性及相关酶活性的影响. 中国农业科学 39, 1220-1227. |
| [10] | 汪开拓, 郑永华, 唐文才, 李廷君, 张卿, 尚海涛 ( 2012). 茉莉酸甲酯处理对葡萄果实NO和H2O2水平及植保素合成的影响. 园艺学报 39, 1559-1566. |
| [11] | 王英珍, 程瑞, 张绍铃, 白彬, 何子顺, 张虎平 ( 2016). 采前茉莉酸甲酯(MeJA)处理对梨果实抗病性的影响. 果树学报 33, 694-700. |
| [12] | 许晴晴, 郜海燕, 陈杭君 ( 2014). 茉莉酸甲酯对蓝莓贮藏品质及抗病相关酶活性的影响. 核农学报 28, 1226-1231. |
| [13] | 张芮 ( 2015). 苹果MdWRKY33基因在轮纹病抗性形成中的作用机制研究. 博士论文. 泰安: 山东农业大学. pp. 70-73. |
| [14] | Chen XK, Zhang JY, Zhang Z, Du XL, Du BB, Qu SC ( 2012). Overexpressing MhNPR1 in transgenic Fuji apples enhances resistance to apple powdery mildew. Mol Biol Rep 39, 8083-8089. |
| [15] | Langmead B, Trapnell C, Pop M, Salzberg SL ( 2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. |
| [16] | Li H, Durbin R ( 2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-1760. |
| [17] | Sun X, Matus JT, Wong DCJ, Wang Z, Chai F, Zhang L, FangT, Zhao L, Wang Y, Han Y, Wang Q, Li S, Liang Z, Xin H ( 2018). The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J Exp Bot 69, 1749-1764. |
| [18] | Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen ZT, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu CJ, Chu VT, King ST, Vick J, Tao QZ, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagné D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouzé P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R ( 2010). The genome of the domesticated apple ( Malus × domestica Borkh.). Nat Genet 42, 833-839. |
| [19] | Xue YK, Shui GH, Wenk MR ( 2014). TPS1 drug design for rice blast disease in Magnaporthe oryzae. Springer Plus 3, 18. |
| [20] | Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J ( 2006). WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34, W293-W297. |
| [21] | Zhang HY, Ma LC, Turner M, Xu HX, Dong Y, Jiang S ( 2009). Methyl jasmonate enhances biocontrol efficacy of Rhodotorula glutinis to postharvest blue mold decay of pears. Food Chem 117, 621-626. |
| [22] | Zhang Y, Shi XP, Li BH, Zhang QM, Liang WX, Wang CX ( 2016). Salicylic acid confers enhanced resistance to Glomerella leaf spot in apple. Plant Physiol Biochem 106, 64-72. |
| [1] |
Liang Ma, Yongqing Yang, Yan Guo.
“Next-generation Green Revolution” Genes: Toward New “Climate-Smart” Crop Breeding [J]. Chinese Bulletin of Botany, 2025, 60(4): 489-498. |
| [2] | Xiangge Zhang, Chen Chen, Shan Cheng, Chunxin Li, Yajing Zhu, Xinran Xu, Huiwei Wang. Identification and Analysis of Tuber-specific Expression Genes in Cyperus esculentus [J]. Chinese Bulletin of Botany, 2025, 60(1): 33-48. |
| [3] | Yuying Zhou, Hui Chen, Simu Liu. Research Progress on Auxin Responsive Non-canonical Aux/IAA Proteins in Plants [J]. Chinese Bulletin of Botany, 2024, 59(4): 651-658. |
| [4] | Jianmin Zhou. A Combat Vehicle with a Smart Brake [J]. Chinese Bulletin of Botany, 2024, 59(3): 343-346. |
| [5] | JIANG Hai-Gang, ZENG Yun-Hong, TANG Hua-Xin, LIU Wei, LI Jie-Lin, HE Guo-Hua, QIN Hai-Yan, WANG Li-Chao, Victor RESCO de DIOS, YAO Yin-An. Rhythmic regulation of carbon fixation and water dissipation in three mosses [J]. Chin J Plant Ecol, 2023, 47(7): 988-997. |
| [6] | Nan Wu, Lei Qin, Kan Cui, Haiou Li, Zhongsong Liu, Shitou Xia. Cloning of Brassica napus EXA1 Gene and Its Regulation on Plant Disease Resistance [J]. Chinese Bulletin of Botany, 2023, 58(3): 385-393. |
| [7] | BAI Xue, LI Yu-Jing, JING Xiu-Qing, ZHAO Xiao-Dong, CHANG Sha-Sha, JING Tao-Yu, LIU Jin-Ru, ZHAO Peng-Yu. Response mechanisms of millet and its rhizosphere soil microbial communities to chromium stress [J]. Chin J Plant Ecol, 2023, 47(3): 418-433. |
| [8] | LU Chen-Xi, XU Man, SHI Xue-Jin, ZHAO Cheng, TAO Ze, LI Min, SI Bing-Cheng. Effects of different water isotope input methods based on Bayesian model MixSIAR on water uptake characteristic analysis results in apple orchards [J]. Chin J Plant Ecol, 2023, 47(2): 238-248. |
| [9] | Jiman Li, Nan Jin, Maogang Xu, Jusong Huo, Xiaoyun Chen, Feng Hu, Manqiang Liu. Effects of earthworm on tomato resistance under different drought levels [J]. Biodiv Sci, 2022, 30(7): 21488-. |
| [10] | Li Yue, Hu Desheng, Tan Jinfang, Mei Hao, Wang Yi, Li Hui, Li Fang, Han Yanlai. Chaetomium uniseriatum Promotes Maize Growth by Accelerating Straw Degradation and Regulating the Expression of Hormone Responsive Genes [J]. Chinese Bulletin of Botany, 2022, 57(4): 422-433. |
| [11] | Lixia Jia, Yanhua Qi. Advances in the Regulation of Rice (Oryza sativa) Grain Shape by Auxin Metabolism, Transport and Signal Transduction [J]. Chinese Bulletin of Botany, 2022, 57(3): 263-275. |
| [12] | Tiantian Zhi, Zhou Zhou, Chengyun Han, Chunmei Ren. PAD4 Mutation Accelerating Programmed Cell Death in Arabidopsis thaliana Tyrosine Degradation Deficient Mutant sscd1 [J]. Chinese Bulletin of Botany, 2022, 57(3): 288-298. |
| [13] | Ting Wang, Zengqiang Xia, Jiangping Shu, Jiao Zhang, Meina Wang, Jianbing Chen, Kanglin Wang, Jianying Xiang, Yuehong Yan. Dating whole-genome duplication reveals the evolutionary retardation of Angiopteris [J]. Biodiv Sci, 2021, 29(6): 722-734. |
| [14] | Jian-Min Zhou. A Ca2+-ROS Signaling Axis in Rice Provides Clues to Rice-pathogen Coevolution and Crop Improvements [J]. Chinese Bulletin of Botany, 2021, 56(5): 513-515. |
| [15] | Tiantian Shi, Ying Gao, Huan Wang, Jun Liu. Nucleo-cytoplasmic Transport and Transport Receptors in Plant Disease Resistance Defense Response [J]. Chinese Bulletin of Botany, 2021, 56(4): 480-487. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||