

植物学报 ›› 2017, Vol. 52 ›› Issue (2): 179-187.DOI: 10.11983/CBB16041 cstr: 32102.14.CBB16041
收稿日期:2016-03-08
接受日期:2016-08-08
出版日期:2017-03-01
发布日期:2017-04-05
通讯作者:
包颖
作者简介:# 共同第一作者
基金资助:
Wang Qian, Sun Wenjing, Bao Ying*( )
)
Received:2016-03-08
Accepted:2016-08-08
Online:2017-03-01
Published:2017-04-05
Contact:
Bao Ying
About author:# Co-first authors
摘要: 为全面理解植物颗粒结合淀粉合酶(GBSS)基因在植物中的进化模式并重建其进化历史, 利用20种陆生植物和2种藻类植物的基因组数据, 通过生物信息学手段, 深入挖掘和分析植物类群基因组中GBSS基因家族的构成和基因特点, 推测其可能的扩增和丢失规律。结果共识别42条同源序列。系统发育和进化分析表明, GBSS基因起源古老, 可能在所有绿色植物的祖先中就已经出现, 之后在进化过程中不断发生谱系的特异扩张和拷贝丢失, 并最终通过功能分化的形式在植物类群中被固定。
王倩, 孙文静, 包颖. 植物颗粒结合淀粉合酶GBSS基因家族的进化. 植物学报, 2017, 52(2): 179-187.
Wang Qian, Sun Wenjing, Bao Ying. Evolutionary Pattern of the GBSS Gene Family in Plants. Chinese Bulletin of Botany, 2017, 52(2): 179-187.
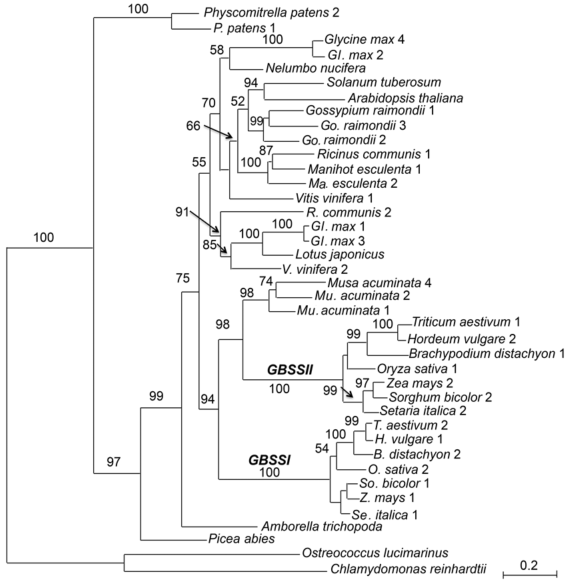
图1 基于22种植物GBSS基因氨基酸序列构建的最大似然系统发育树分支上的数值代表大于50%的最大似然分析的靴带支持率。种名后数字代表种内的不同基因位点, 对这些位点的细节描述见表1。
Figure 1 The maximum likelihood tree of the granule-bound starch synthase gene family based on amino acid sequences of 22 plant speciesNumbers above the branches indicate bootstrap values above 50%. Numbers following species names indicate different gene loci as listed in Table 1.
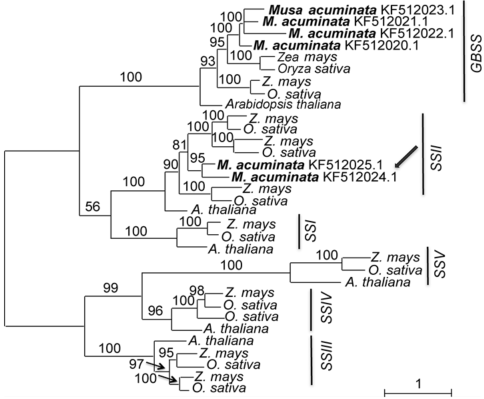
图2 基于4种植物淀粉合酶基因的氨基酸同源序列构建的最大似然树分支上的数值代表大于50%的最大似然分析的靴带支持率。箭头示2条香蕉基因实为SSII而非GBSS。
Figure 2 Maximum likelihood tree of 4 species based on amino acid homologous sequences of the starch synthase genesNumbers above the branches indicate bootstrap values above 50%. Two genes of Musa acuminata indicated by arrow should be SSII rather than GBSS.
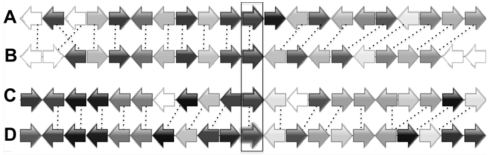
图3 大豆4个GBSS同源基因位点在染色体片段的同线性排列(A) 位点GM10G31540所在染色体片段; (B) 位点GM20- G36040所在染色体片段; (C) 位点GM16G02110所在染色体片段; (D) 位点GM07G05580所在染色体片段。箭头代表不同的基因及其在基因组上的排列方向。同源基因采用相同的颜色。图中框格标注的为4个GBSS基因。
Figure 3 Synteny alignment of the chromosome regions with 4 GBSS homologous genes in Glycine max(A) The chromosome region includes locus GM10G31540; (B) The chromosome region includes locus GM20G36040; (C) The chromosome region includes locus GM16G02110; (D) The chromosome region includes locus GM07G05580. Arrows indicate the direction of genes’ transcription. Homologous genes are shown in same colors. Four GBSS genes are in the frame.
| [1] | 包颖, 杜家潇, 景翔, 徐思 (2015). 药用野生稻叶中淀粉合成酶基因家族的序列分化和特异表达. 植物学报50, 683-690. |
| [2] | Ahuja G, Jaiswal S, Hucl P, Chibbar RN (2014). Wheat genome specific granule-bound starch synthase I differentially influence grain starch synthesis.Carbohydr Polym 114, 87-94. |
| [3] | Alison MS (2012). Starch in the Arabidopsis plant.Starch 64, 421-434. |
| [4] | Ball S, Colleoni C, Cenci U, Raj JN, Tirtiaux C (2011). The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis.J Exp Bot 62, 1775-1801. |
| [5] | Baranov Iu O, Slishchuk HI, Volkova NE, SyvolapIu M (2014). Bioinformatic analysis of maize granule-bound starch synthase gene.Tsitol Genet 48, 18-23. |
| [6] | Criscuolo A (2011). morePhyML: improving the phylogenetic tree space exploration with PhyML 3.Mol Phylogenet Evol 61, 944-948. |
| [7] | Deschamps P, Moreau H, Worden AZ, Dauvillee D, Ball SG (2008). Early gene duplication within chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts.Genetics 178, 2373-2387. |
| [8] | Dian W, Jiang H, Chen Q, Liu F, Wu P (2003). Cloning and characterization of the granule-bound starch synthase II gene in rice: gene expression is regulated by the nitrogen level, sugar and circadian rhythm.Planta 218, 261-268. |
| [9] | Fasahat P, Rahman S, Ratnam W (2014). Genetic controls on starch amylose content in wheat and rice grains.J Genet 93, 279-292. |
| [10] | Fulton DC, Edwards A, Pilling E, Robinson HL, Fahy B, Seale R, Kato L, Donald AM, Geigenberger P, Martin C, Smith AM (2002). Role of granule-bound starch synthase in determination of amylopectin structure and starch granule morphology in potato.J Biol Chem 277, 10834-10841. |
| [11] | Guzman C, Alvarez JB (2015). Wheat waxy proteins: polymorphism, molecular characterization and effects on starch properties.Theor Appl Genet 9, 1049-1060. |
| [12] | Hirose T, Hashida Y, Aoki N, Okamura M, Yonekura M, Ohto C, Terao T, Ohsugi R (2014). Analysis of gene- disruption mutants of a sucrose phosphate synthase gene in rice,OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant Sci 225, 102-106. |
| [13] | Hoai TT, Matsusaka H, Toyosawa Y, Suu TD, Satoh H, Kumamaru T (2014). Influence of single-nucleotide poly- morphisms in the gene encoding granule-bound starch synthase I on amylose content in Vietnamese rice cultivars.Breed Sci 64, 142-148. |
| [14] | Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y (2010). Starch biosynthesis in cereal endosperm.Plant Physiol Biochem 48, 383-392. |
| [15] | Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007). Clustal W and Clustal X version 2.0.Bioinformatics 23, 2947-2948. |
| [16] | Miao H, Sun P, Liu W, Xu B, Jin Z (2014). Identification of genes encoding granule-bound starch synthase involved in amylose metabolism in banana fruit.PLoS One 9, e88077. |
| [17] | Ohdan T, Francisco PB, Sawada TJ, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot 56, 3229-3244. |
| [18] | Orzechowski S (2008). Starch metabolism in leaves.Acta Biochim Pol 55, 435-445. |
| [19] | Patron NJ, Smith AM, Fahy BF, Hylton CM, Naldrett MJ, Rossnagel BG, Denyer K (2002). The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5'-non-coding region.Plant Physiol 130, 190-198. |
| [20] | Tsai CY (1974). The function of the waxy locus in starch synthesis in maize endosperm.Biochem Genet 11, 83-96. |
| [21] | Vrinten PL, Nakamura T (2000). Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues.Plant Physiol 122, 255-264. |
| [22] | Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench.Bioinformatics 25, 1189-1191. |
| [23] | Yan HB, Pan XX, Jiang HW, Wu GJ (2009). Comparison of the starch synthesis genes between maize and rice: copies, chromosome location and expression divergence.Theor Appl Genet 119, 815-825. |
| [24] | Zhu L, Gu M, Meng X, Cheung SC, Yu H, Huang J, Sun Y, Shi Y, Liu Q (2012). High-amylose rice improves indices of animal health in normal and diabetic rats.Plant Biotechnol J 10, 353-362. |
| [1] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | 卢晓强, 董姗姗, 马月, 徐徐, 邱凤, 臧明月, 万雅琼, 李孪鑫, 于赐刚, 刘燕. 前沿技术在生物多样性研究中的应用现状、挑战与展望[J]. 生物多样性, 2025, 33(4): 24440-. |
| [3] | 崔娟, 于晓玉, 于跃娇, 梁铖玮, 孙健, 陈温福. 影响中国东北和日本粳稻食味品质差异的质构因素及其遗传基础解析[J]. 植物学报, 2025, 60(4): 1-0. |
| [4] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [5] | 张如礼, 李德铢, 张玉霄. 短穗竹居群遗传结构及气候适应性分析[J]. 植物学报, 2025, 60(3): 407-424. |
| [6] | 林珍, 向家宝, 蔡何佳奕, 高贝, 杨金涛, 李俊毅, 周青松, 黄晓磊, 邓鋆. 七种半翅目昆虫线粒体基因组数据[J]. 生物多样性, 2025, 33(2): 24434-. |
| [7] | 曹东, 李焕龙, 彭扬, 魏存争. 植物基因组大小与性状关系的研究进展[J]. 生物多样性, 2025, 33(2): 24192-. |
| [8] | 夏琳凤, 李瑞, 王海政, 冯大领, 王春阳. 轮藻门植物基因组学研究进展[J]. 植物学报, 2025, 60(2): 271-282. |
| [9] | 孙亚君. 何谓高等或低等生物——澄清《物种起源》所蕴含的生物等级性的涵义及其成立性[J]. 生物多样性, 2025, 33(1): 24394-. |
| [10] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [11] | 姚祥坦, 张心怡, 陈阳, 袁晔, 程旺大, 王天瑞, 邱英雄. 基于基因组重测序揭示栽培欧菱遗传多样性及‘南湖菱’的起源驯化历史[J]. 生物多样性, 2024, 32(9): 24212-. |
| [12] | 张强, 赵振宇, 李平华. 基因编辑技术在玉米中的研究进展[J]. 植物学报, 2024, 59(6): 978-998. |
| [13] | 李园, 范开建, 安泰, 李聪, 蒋俊霞, 牛皓, 曾伟伟, 衡燕芳, 李虎, 付俊杰, 李慧慧, 黎亮. 玉米自然群体自交系农艺性状的多环境全基因组预测初探[J]. 植物学报, 2024, 59(6): 1041-1053. |
| [14] | 陈文娜, 李良涛, 周璐, 姚纲. 太行山近期隆升促进太行花属(蔷薇科)谱系分化[J]. 植物学报, 2024, 59(5): 763-773. |
| [15] | 艾妍雨, 胡海霞, 沈婷, 莫雨轩, 杞金华, 宋亮. 附生维管植物多样性及其与宿主特征的相关性: 以哀牢山中山湿性常绿阔叶林为例[J]. 生物多样性, 2024, 32(5): 24072-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||