

植物学报 ›› 2018, Vol. 53 ›› Issue (4): 542-555.DOI: 10.11983/CBB18080 cstr: 32102.14.CBB18080
收稿日期:2018-03-30
接受日期:2018-05-02
出版日期:2018-07-01
发布日期:2018-09-11
通讯作者:
周玉萍
作者简介:† 共同第一作者。
基金资助:
Wu Jinghui, Xie Chuping, Tian Changen, Zhou Yuping*( )
)
Received:2018-03-30
Accepted:2018-05-02
Online:2018-07-01
Published:2018-09-11
Contact:
Zhou Yuping
About author:† These authors contributed equally to this paper
摘要: 脱落酸(ABA)是调控种子休眠和萌发过程的主要植物激素。种子内源ABA含量和种胚对ABA敏感性共同调控种子休眠和萌发过程, 确保植物种子以休眠状态在逆境中保持其自身繁衍能力, 并在适宜的环境下启动萌发程序。种子ABA合成代谢和ABA信号转导途径涉及许多重要基因家族, 它们通过复杂的调控网络精确地控制着种胚发生、种子成熟、休眠及萌发进程。该文对ABA调控种子休眠和萌发的分子机制最新研究进展进行综述, 并展望了今后的研究方向。
伍静辉, 谢楚萍, 田长恩, 周玉萍. 脱落酸调控种子休眠和萌发的分子机制. 植物学报, 2018, 53(4): 542-555.
Wu Jinghui, Xie Chuping, Tian Changen, Zhou Yuping. Molecular Mechanism of Abscisic Acid Regulation During Seed Dormancy and Germination. Chinese Bulletin of Botany, 2018, 53(4): 542-555.
| 基因名称 | 突变体休眠能力 | 基本功能 | 参考文献 |
|---|---|---|---|
| NCED6/9 | 变化不显著 | ABA合成途径关键代谢酶 | Lefebvre et al., 2006 |
| CYP707A1/2 | 增强 | ABA代谢途径关键代谢酶, 其表达受ABI4调控 | Kushiro et al., 2004; Okamoto et al., 2006; Shu et al., 2013 |
| ABI3 | 降低 | 促进种子发育, 抑制种子萌发 | Raz et al., 2001; Dekkers et al., 2016 |
| FUS3 | 降低 | 促进种子发育, 增强NCED6和NCED9的表达 | Raz et al., 2001; Tiedemann et al., 2008; Yamamoto et al., 2010 |
| LEC2 | 降低 | 促进种子发育, 促进LEC1、FUC3和DOG1的表达 | Raz et al., 2001; Braybrook et al., 2006; Stone et al., 2008 |
| LEC1/L1L | 降低 | 促进种子发育, 激活ABI3、FUS3和LEC2的表达 | Raz et al., 2001; Kagaya et al., 2005; To et al., 2006 |
| DEP | 降低 | 促进ABI3的表达 | Barrero et al., 2010 |
| DOG1 | 降低 | 种子休眠正向调控因子 | Née et al., 2017 |
| ABI4 | 降低 | 促进种子发育, 是ABA/GA的关键调控因子 | Shu et al., 2013 |
| SPT | Col背景中增强; Ler背景中降低 | 调控ABI4表达, 在Col和Ler背景下功能相反 | Belmonte et al., 2013; Vaistij et al., 2013 |
| CHO1 | 降低 | 促进ABA合成, 抑制ABA降解 | Yano et al., 2009 |
| MYB96 | 降低 | 促进种子休眠, 抑制种子萌发 | Lee et al., 2015a, 2015b |
| ABI5 | 变化不显著 | 不直接参与种子休眠调控, 但负调控种子萌发 | Piskurewicz et al., 2008; Kanai et al., 2010 |
| bZIP10/25/53 | 降低 | 协同促进种子发育 | Alonso et al., 2009 |
| WRKY41 | 降低 | 促进ABI3转录 | Ding et al., 2014 |
| DAG1 | 降低 | 促进ABA而抑制GA积累, 正调控种子休眠 | Boccaccini et al., 2014, 2016 |
| DOF6 | 未提及 | 与RGL2构成复合体, 共同促进种子初级休眠 | Ravindran et al., 2017 |
| KYP/SUVH4 | 增强 | 抑制DOG1和ABI3转录, 负调控种子休眠 | Zheng et al., 2012 |
| LDL1/LDL2 | 增强 | 抑制DOG1表达, 负调控种子休眠 | Zhao et al., 2015 |
表1 参与种子休眠的关键基因
Table 1 Key genes involved in seed dormancy
| 基因名称 | 突变体休眠能力 | 基本功能 | 参考文献 |
|---|---|---|---|
| NCED6/9 | 变化不显著 | ABA合成途径关键代谢酶 | Lefebvre et al., 2006 |
| CYP707A1/2 | 增强 | ABA代谢途径关键代谢酶, 其表达受ABI4调控 | Kushiro et al., 2004; Okamoto et al., 2006; Shu et al., 2013 |
| ABI3 | 降低 | 促进种子发育, 抑制种子萌发 | Raz et al., 2001; Dekkers et al., 2016 |
| FUS3 | 降低 | 促进种子发育, 增强NCED6和NCED9的表达 | Raz et al., 2001; Tiedemann et al., 2008; Yamamoto et al., 2010 |
| LEC2 | 降低 | 促进种子发育, 促进LEC1、FUC3和DOG1的表达 | Raz et al., 2001; Braybrook et al., 2006; Stone et al., 2008 |
| LEC1/L1L | 降低 | 促进种子发育, 激活ABI3、FUS3和LEC2的表达 | Raz et al., 2001; Kagaya et al., 2005; To et al., 2006 |
| DEP | 降低 | 促进ABI3的表达 | Barrero et al., 2010 |
| DOG1 | 降低 | 种子休眠正向调控因子 | Née et al., 2017 |
| ABI4 | 降低 | 促进种子发育, 是ABA/GA的关键调控因子 | Shu et al., 2013 |
| SPT | Col背景中增强; Ler背景中降低 | 调控ABI4表达, 在Col和Ler背景下功能相反 | Belmonte et al., 2013; Vaistij et al., 2013 |
| CHO1 | 降低 | 促进ABA合成, 抑制ABA降解 | Yano et al., 2009 |
| MYB96 | 降低 | 促进种子休眠, 抑制种子萌发 | Lee et al., 2015a, 2015b |
| ABI5 | 变化不显著 | 不直接参与种子休眠调控, 但负调控种子萌发 | Piskurewicz et al., 2008; Kanai et al., 2010 |
| bZIP10/25/53 | 降低 | 协同促进种子发育 | Alonso et al., 2009 |
| WRKY41 | 降低 | 促进ABI3转录 | Ding et al., 2014 |
| DAG1 | 降低 | 促进ABA而抑制GA积累, 正调控种子休眠 | Boccaccini et al., 2014, 2016 |
| DOF6 | 未提及 | 与RGL2构成复合体, 共同促进种子初级休眠 | Ravindran et al., 2017 |
| KYP/SUVH4 | 增强 | 抑制DOG1和ABI3转录, 负调控种子休眠 | Zheng et al., 2012 |
| LDL1/LDL2 | 增强 | 抑制DOG1表达, 负调控种子休眠 | Zhao et al., 2015 |
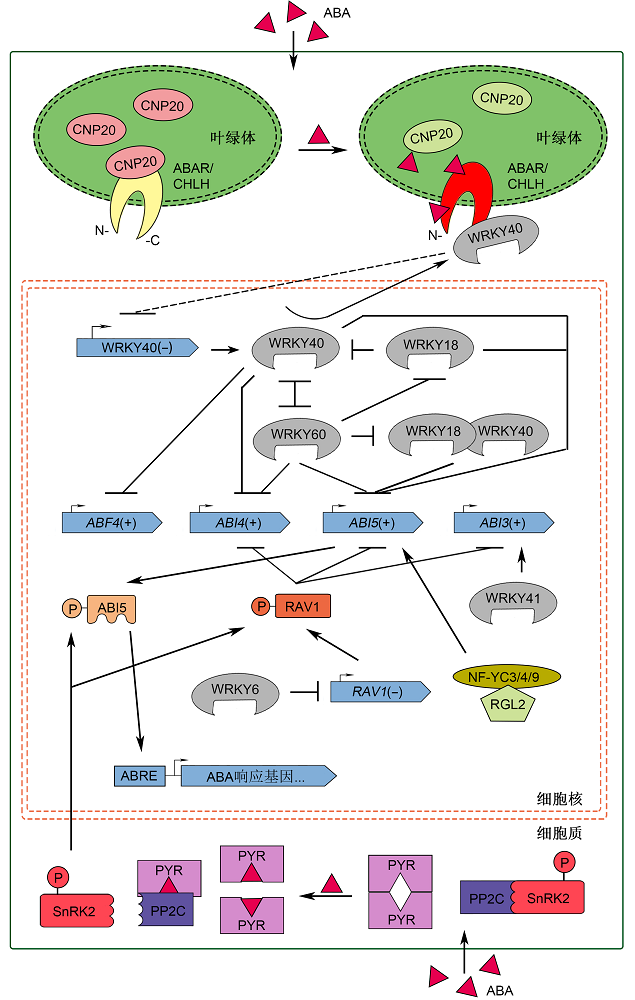
图1 以PYR/PYL/RCAR和ABAR/CHLH为受体的ABA信号转导通路模型细胞质ABA受体PYR/PYL/RCAR在静息状态下以二聚体形式存在, 当与ABA结合后则以单体形式与PP2Cs结合, 解除对SnRK2s的抑制, 磷酸化ABI5及RAV1等转录因子, 激活下游ABA响应基因。ABAR/CHLH是叶绿体膜上的ABA受体, 其C-端和N-端暴露在细胞质中。低浓度ABA时, 叶绿体内伴侣蛋白CPN20结合ABAR, 抑制ABAR与负调控因子WRKY40的相互作用, 同时WRKY40与WRKY18和WRKY60通过直接或间接作用, 抑制ABF4/ABI4/ABI5的表达。高浓度ABA促使WRKY40向细胞质迁移, ABAR的C-端与WRKY40相互作用, 解除WRKY40对ABF4/ABI4/ABI5的转录抑制, 实现ABA生理效应。RAV1通过抑制ABI3/ABI4/ABI5转录来负调控ABA信号; WRKY6、WRKY41和NF-YC-RGL2分别通过下调RAV1的表达, 促进ABI3和ABI5的表达, 它们均正调控ABA信号。图中(+)和(-)分别表示该基因在ABA信号通路中具有正向和反向调控作用。
Figure 1 Models of ABA signaling transduction pathway based on ABA receptors PYR/PYL/RCAR and ABAR/CHLHIntracellular ABA receptor PYR/PYL/RCAR exists as a homodimer in the resting state. Upon binding to ABA, PYR/PYL/RCAR turns into monomer and interacts with PP2Cs to release SnRK2s, which phosphorylates transcription factors like ABI5 and RAV1, to activate downstream ABA responsive genes. ABAR/CHLH is a protein across the chloroplast membrane, whose C- and N- ends are exposed to the cytoplasm. In the absence or low levels of ABA, the interaction of CPN20 with ABAR attenuates the interaction between ABAR and the negative regulator WRKY40, meanwhile WRKY40, WRKY18 and WRKY60 directly or indirectly inhibit the expression of ABF4/ABI4/ABI5. In response to a high level of ABA, WRKY40 is recruited from nucleus to the cytosol. C-terminal part of ABA-bound ABAR interacts with WRKY40 and relieves the suppression of ABF4/ABI4/ABI5 by WRKY40, promoting the physiological effects of ABA. RAV1 negatively regulates the ABA signaling by inhibiting the transcription of ABI3/ABI4/ABI5. WRKY6, WRKY41, and NF-YC-RGL2 play the positive roles in the ABA signaling, by down-regulating the expression of RAV1, promoting the expression of ABI3 and ABI5, respectively. (+) and (-) in this figure denote the positive regulation and the negative regulation in the ABA signaling pathway, respectively.
| 1 | 操春燕, 陈熙, 吴燕 (2017). 脱落酸调控种子萌发和休眠的研究进展. 生命科学 29, 582-588. |
| 2 | 刘一灵, 李振华, 刘仁祥 (2014). 植物激素ABA调控种子发育与萌发的研究进展. 种子 33(10), 47-50. |
| 3 | 曾后清, 张夏俊, 张亚仙, 汪尚, 皮二旭, 王慧中, 杜立群 (2016). 植物类钙调素生理功能的研究进展. 中国科学: 生命科学 46, 705-715. |
| 4 | 张大鹏 (2011). 始于质体/叶绿体的ABA信号通路. 植物学报 46, 361-369. |
| 5 | Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation.Plant Cell 21, 1747-1761. |
| 6 | Barrero JM, Millar AA, Griffiths J, Czechowski T, Scheible WR, Udvardi M, Reid JB, Ross JJ, Jacobsen JV, Gubler F (2010). Gene expression profiling identifies two re- gulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds.Plant J 61, 611-622. |
| 7 | Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed.Proc Natl Acad Sci USA 110, E435-E444. |
| 8 | Bienz M (2006). The PHD finger, a nuclear protein-interac- tion domain.Trends Biochem Sci 31, 35-40. |
| 9 | Boccaccini A, Lorrai R, Ruta V, Frey A, Mercey-Boutet S, Marion-Poll A, Tarkowská D, Strnad M, Costantino P, Vittorioso P (2016). The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels.BMC Plant Biol 16, 198. |
| 10 | Boccaccini A, Santopolo S, Capauto D, Lorrai R, Minutello E, Belcram K, Palauqui JC, Costantino P, Vittorioso P (2014). Independent and interactive effects of DOF affecting germination 1 (DAG1) and the Della proteins GA insensitive (GAI) and repressor of ga1-3 (RGA) in embryo development and seed germination. BMC Plant Biol 14, 200. |
| 11 | Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006). Genes directly regulated by LEAFY COTYLEDON 2 provide insight into the control of embryo maturation and somatic embryogenesis.Proc Natl Acad Sci USA 103, 3468-3473. |
| 12 | Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006). Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism.Plant J 46, 805-822. |
| 13 | Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002). Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30, 373-383. |
| 14 | Chen H, Lai ZB, Shi JW, Xiao Y, Chen ZX, Xu XP (2010). Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress.BMC Plant Biol 10, 281. |
| 15 | Chen HY, Hsieh EJ, Cheng MC, Chen CY, Hwang SY, Lin TP (2016). ORA47 (octadecanoid-responsive AP2/ERF- domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through bin- ding to a novel cis-element. New Phytol 211, 599-613. |
| 16 | Dekkers BJW, He HZ, Hanson J, Willems LAJ, Jamar DCL, Cueff G, Rajjou L, Hilhorst HWM, Bentsink L (2016). The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J 85, 451-465. |
| 17 | Ding ZJ, Yan JY, Li GX, Wu ZC, Zhang SQ, Zheng SJ (2014). WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J 79, 810-823. |
| 18 | Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J 80, 654-668. |
| 19 | Finch-Savage WE, Leubner-Metzger G (2006). Seed dormancy and the control of germination.New Phytol 171, 501-523. |
| 20 | Finkelstein RR, Gampala SSL, Rock CD (2002). Abscisic acid signaling in seeds and seedlings.Plant Cell 14, S15-S45. |
| 21 | Finkelstein RR, Lynch TJ (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599-610. |
| 22 | Footitt S, Müller K, Kermode AR, Finch-Savage WE (2015). Seed dormancy cycling in Arabidopsis: chromatin remodelling and regulation of DOG1 in response to seasonal environmental signals.Plant J 81, 413-425. |
| 23 | Frey A, Effroy D, Lefebvre V, Seo M, Perreau F, Berger A, Sechet J, To A, North HM, Marion-Poll A (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members.Plant J 70, 501-512. |
| 24 | Fujii H, Verslues PE, Zhu JK (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis.Plant Cell 19, 485-494. |
| 25 | Fujii H, Zhu JK (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress.Proc Natl Acad Sci USA 106, 8380-8385. |
| 26 | Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C (2007). Combined networks regulating seed maturation.Trends Plant Sci 12, 294-300. |
| 27 | Hauser F, Waadt R, Schroeder JI (2011). Evolution of abscisic acid synthesis and signaling mechanisms.Curr Biol 21, R346-R355. |
| 28 | Holdsworth MJ, Bentsink L, Soppe WJJ (2008). Molecular networks regulating Arabidopsis seed maturation, after- ripening, dormancy and germination.New Phytol 179, 33-54. |
| 29 | Huang MK, Hu YL, Liu X, Li YG, Hou XL (2015). Arabidopsis LEAFY COTYLEDON 1 mediates postembryonic development via interacting with PHYTOCHROME-INTER- ACTING FACTOR 4.Plant Cell 27, 3099-3111. |
| 30 | Huang Y, Feng CZ, Ye Q, Wu WH, Chen YF (2016). Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development.PLoS Genet 12, e1005833. |
| 31 | Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Sch- roeder JI (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions.Genes Dev 24, 1695-1708. |
| 32 | Je J, Chen H, Song C, Lim CO (2014). Arabidopsis DREB2C modulates ABA biosynthesis during germination.Biochem Biophys Res Commun 452, 91-98. |
| 33 | Jia HY, Suzuki M, McCarty DR (2014). Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks.Wiley Interdiscip Rev Dev Biol 3, 135-145. |
| 34 | Jiang WB, Yu DQ (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid.BMC Plant Biol 9, 96. |
| 35 | Junker A, Mönke G, Rutten T, Keilwagen J, Seifert M, Thi TM, Renou JP, Balzergue S, Viehöver P, Hähnel U, Ludwig-Müller J, Altschmied L, Conrad U, Weisshaar B, Bäumlein H (2012). Elongation-related functions of LEAFY COTYLEDON 1 during the development of Arabidopsis thaliana. Plant J 71, 427-442. |
| 36 | Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005). LEAFY COTYLEDON 1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE 3. Plant Cell Physiol 46, 399-406. |
| 37 | Kanai M, Nishimura M, Hayashi M (2010). A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J 62, 936-947. |
| 38 | Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, Seo M (2010). Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions.Plant Cell Physiol 51, 1988-2001. |
| 39 | Karssen CM, Brinkhorst-van der Swan DLC, Breekland AE, Koornneef M (1983). Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157, 158-165. |
| 40 | Kondhare KR, Hedden P, Kettlewell PS, Farrell AD, Monaghan JM (2014). Quantifying the impact of exogenous abscisic acid and gibberellins on pre-maturity α- amylase formation in developing wheat grains.Sci Rep 4, 5355. |
| 41 | Koornneef M, Karssen CM (1994). Seed dormancy and germination. In: Meyerowitz EM, Somerville CR, eds. Arabidopsis. New York: Cold Spring Harbor Laboratory Press. pp. 313-334. |
| 42 | Kumimoto RW, Siriwardana CL, Gayler KK, Risinger JR, Siefers N, Holt BF 3rd (2013). NUCLEAR FACTOR Y transcription factors have both opposing and additive roles in ABA-mediated seed germination.PLoS One 8, e59481. |
| 43 | Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism.EMBO J 23, 1647-1656. |
| 44 | Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development.Plant Cell 15, 5-18. |
| 45 | Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J (2003). Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2.J Biol Chem 278, 21003-21011. |
| 46 | Lee HG, Lee K, Seo PJ (2015a). The Arabidopsis MYB96 transcription factor plays a role in seed dormancy.Plant Mol Biol 87, 371-381. |
| 47 | Lee K, Lee HG, Yoon S, Kim HU, Seo PJ (2015b). The Arabidopsis MYB96 transcription factor is a positive regulator of ABSCISIC ACID-INSENSITIVE 4 in the control of seed germination. Plant Physiol 168, 677-689. |
| 48 | Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45, 309-319. |
| 49 | Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH (2007). Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol 143, 745-758. |
| 50 | Liu X, Hu P, Huang M, Tang Y, Li Y, Li L, Hou X (2016). The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis.Nat Com- mun 7, 12768. |
| 51 | Liu YX, Geyer R, van Zanten M, Carles A, Li Y, Hörold A, van Nocker S, Soppe WJJ (2011). Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS One 6, e22241. |
| 52 | Liu ZQ, Yan L, Wu Z, Mei C, Lu K, Yu YT, Liang S, Zhang XF, Wang XF, Zhang DP (2012). Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J Exp Bot 63, 6371-6392. |
| 53 | Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination.Plant J 32, 317-328. |
| 54 | Lotan T, Ohto MA, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998). Arabidopsis LEAFY COTYLEDON 1 is sufficient to induce embryo development in vegetative cells.Cell 93, 1195-1205. |
| 55 | Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors.Science 324, 1064-1068. |
| 56 | Midhat U, Ting MKY, Teresinski HJ, Snedden WA (2018). The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis.Plant Mol Biol 96, 375-392. |
| 57 | Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, Altschmied L (2012). Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40, 8240-8254. |
| 58 | Müller K, Carstens AC, Linkies A, Torres MA, Leubner-Metzger G (2009). The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol 184, 885-897. |
| 59 | Nakamura S, Lynch TJ, Finkelstein RR (2001). Physical interactions between ABA response loci of Arabidopsis.Plant J 26, 627-635. |
| 60 | Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2009). Three Arabidopsis SnRK2 protein kinases, SRK- 2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK- 2.3, involved in ABA signaling are essential for the control of seed development and dormancy.Plant Cell Physiol 50, 1345-1363. |
| 61 | Née G, Kramer K, Nakabayashi K, Yuan BJ, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ (2017). DELAY OF GERMINATION 1 requires PP2C phosphatases of the ABA signaling pathway to control seed dormancy.Nat Commun 8, 72. |
| 62 | Nonogaki M, Sall K, Nambara E, Nonogaki H (2014). Amplification of ABA biosynthesis and signaling through a positive feedback mechanism in seeds.Plant J 78, 527-539. |
| 63 | Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination.Plant Cell 15, 1591-1604. |
| 64 | Oh E, Yamaguchi S, Hu JH, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192-1208. |
| 65 | Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis.Plant Phy- siol 141, 97-107. |
| 66 | Papaefthimiou D, Likotrafiti E, Kapazoglou A, Bladenopoulos K, Tsaftaris A (2010). Epigenetic chromatin modifiers in barley: III. Isolation and characterization of the barley GNAT-MYST family of histone acetyltransferases and responses to exogenous ABA.Plant Physiol Biochem 48, 98-107. |
| 67 | Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins.Science 324, 1068-1071. |
| 68 | Perry J, Zhao YD (2003). The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants.Trends Biochem Sci 28, 576-580. |
| 69 | Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity.Plant Cell 20, 2729-2745. |
| 70 | Preston J, Tatematsu K, Kanno Y, Hobo T, Kimura M, Jikumaru Y, Yano R, Kamiya Y, Nambara E (2009). Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: a comparative study on dormant and non-dormant accessions. Plant Cell Physiol 50, 1786-1800. |
| 71 | Ravindran P, Verma V, Stamm P, Kumar PP (2017). A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol Plant 10, 1307-1320. |
| 72 | Raz V, Bergervoet JH, Koornneef M (2001). Sequential steps for developmental arrest in Arabidopsis seeds.Development 128, 243-252. |
| 73 | Ren XZ, Chen ZZ, Liu Y, Zhang HR, Zhang M, Liu Q, Hong XH, Zhu JK, Gong ZZ (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis.Plant J 63, 417-429. |
| 74 | Roscoe TT, Guilleminot J, Bessoule JJ, Berger F, Devic M (2015). Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE 3, FUSCA 3 and LEAFY COTYLEDON 2 in Arabidopsis. Plant Cell Physiol 56, 1215-1228. |
| 75 | Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995). Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol Biol 29, 691-702. |
| 76 | Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez- Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signaling.Plant J 37, 354-369. |
| 77 | Santopolo S, Boccaccini A, Lorrai R, Ruta V, Capauto D, Minutello E, Serino G, Costantino P, Vittorioso P (2015). DOF AFFECTING GERMINATION 2 is a positive regulator of light-mediated seed germination and is repressed by DOF AFFECTING GERMINATION 1.BMC Plant Biol 15, 72. |
| 78 | Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, Zhang XF, Zhao R, Sun HL, Liu R, Yu YT, Zhang DP (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition.Plant Cell 22, 1909-1935. |
| 79 | Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, Fan RC, Xu YH, Zhang DP (2006). The Mg-chelatase H subunit is an abscisic acid receptor.Nature 443, 823-826. |
| 80 | Shu K, Liu XD, Xie Q, He ZH (2016). Two faces of one seed: hormonal regulation of dormancy and germination.Mol Plant 9, 34-45. |
| 81 | Shu K, Zhang HW, Wang SF, Chen ML, Wu YR, Tang SY, Liu CY, Feng YQ, Cao XF, Xie Q (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis.PLoS Genet 9, e1003577. |
| 82 | Shu K, Zhou WG, Yang WY (2018). APETALA 2-domain- containing transcription factors: focusing on abscisic acid and gibberellins antagonism.New Phytol 217, 977-983. |
| 83 | Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ (2008). Arabidopsis LEAFY COTYLEDON 2 induces maturation traits and auxin activity: implications for somatic embryogenesis.Proc Natl Acad Sci USA 105, 3151-3156. |
| 84 | Suzuki M, Wang HHY, McCarty DR (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143, 902-911. |
| 85 | Tiedemann J, Rutten T, Mönke G, Vorwieger A, Rolletschek H, Meissner D, Milkowski C, Petereck S, Mock HP, Zank T, Bäumlein H (2008). Dissection of a complex seed phenotype: novel insights of FUSCA3 regulated developmental processes. Dev Biol 317, 1-12. |
| 86 | To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006). A network of local and redundant gene regulation governs Arabidopsis seed maturation.Plant Cell 18, 1642-1651. |
| 87 | Vaistij FE, Gan YB, Penfield S, Gilday AD, Dave A, He ZS, Josse EM, Choi G, Halliday KJ, Graham IA (2013). Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA.Proc Natl Acad Sci USA 110, 10866-10871. |
| 88 | Virdi AS, Singh S, Singh P (2015). Abiotic stress responses in plants: roles of calmodulin-regulated proteins.Front Plant Sci 6, 809. |
| 89 | Wang FF, Perry SE (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development.Plant Physiol 161, 1251-1264. |
| 90 | Weitbrecht K, Müller K, Leubner-Metzger G (2011). First off the mark: early seed germination.J Exp Bot 62, 3289-3309. |
| 91 | Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, Hattori T (2010). Diverse roles and mechanisms of gene regu- lation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis.Plant Cell Physiol 51, 2031-2046. |
| 92 | Yano R, Kanno Y, Jikumaru Y, Nakabayashi K, Kamiya Y, Nambara E (2009). CHOTTO1, a putative double APETA- LA2 repeat transcription factor, is involved in abscisic acid- mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis.Plant Physiol 151, 641-654. |
| 93 | Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006). ABA-hyper- sensitive germination 3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phospha- tase 2Cs. Plant Physiol 140, 115-126. |
| 94 | Zhang DP (2014). Abscisic Acid: Metabolism, Transport and Signaling. Dordrecht: Springer. pp. 33-35. |
| 95 | Zhang XF, Jiang T, Wu Z, Du SY, Yu YT, Jiang SC, Lu K, Feng XJ, Wang XF, Zhang DP (2013). Cochaperonin CPN20 negatively regulates abscisic acid signaling in Ara- bidopsis.Plant Mol Biol 83, 205-218. |
| 96 | Zhang XF, Jiang T, Yu YT, Wu Z, Jiang SC, Lu K, Feng XJ, Liang S, Lu YF, Wang XF, Zhang DP (2014). Arabidopsis co-chaperonin CPN20 antagonizes Mg-chelatase H subunit to derepress ABA-responsive WRKY40 transcription repressor.Sci China Life Sci 57, 11-21. |
| 97 | Zhao ML, Yang SG, Liu XC, Wu KQ (2015). Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes.Front Plant Sci 6, 159. |
| 98 | Zheng J, Chen FY, Wang Z, Cao H, Li XY, Deng X, Soppe WJJ, Li Y, Liu YX (2012). A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy.New Phytol 193, 605-616. |
| [1] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | 孙龙, 李文博, 娄虎, 于澄, 韩宇, 胡同欣. 火干扰对兴安落叶松种子萌发的影响[J]. 植物生态学报, 2024, 48(6): 770-779. |
| [3] | 罗燕, 刘奇源, 吕元兵, 吴越, 田耀宇, 安田, 李振华. 拟南芥光敏色素突变体种子萌发的光温敏感性[J]. 植物学报, 2024, 59(5): 752-762. |
| [4] | 袁涵, 钟爱文, 刘送平, 彭焱松, 徐磊. 水毛花种子萌发特性的差异及休眠解除方法[J]. 植物生态学报, 2024, 48(5): 638-650. |
| [5] | 朱晓博, 董张, 祝梦瑾, 胡晋, 林程, 陈敏, 关亚静. 重要的种子储存物质长寿命mRNA[J]. 植物学报, 2024, 59(3): 355-372. |
| [6] | 蔡淑钰, 刘建新, 王国夫, 吴丽元, 宋江平. 褪黑素促进镉胁迫下番茄种子萌发的调控机理[J]. 植物学报, 2023, 58(5): 720-732. |
| [7] | 刘晓龙, 季平, 杨洪涛, 丁永电, 付佳玲, 梁江霞, 余聪聪. 脱落酸对水稻抽穗开花期高温胁迫的诱抗效应[J]. 植物学报, 2022, 57(5): 596-610. |
| [8] | 周玉萍, 颜嘉豪, 田长恩. 保卫细胞中ABA信号调控机制研究进展[J]. 植物学报, 2022, 57(5): 684-696. |
| [9] | 张一弓, 张怡, 阿依白合热木·木台力甫, 张道远. 异源过表达齿肋赤藓ScABI3基因改变拟南芥气孔表型并提高抗旱性[J]. 植物学报, 2021, 56(4): 414-421. |
| [10] | 宋松泉, 刘军, 杨华, 张文虎, 张琪, 高家东. 细胞分裂素调控种子发育、休眠与萌发的研究进展[J]. 植物学报, 2021, 56(2): 218-231. |
| [11] | 李绍阳, 马红媛, 赵丹丹, 马梦谣, 亓雯雯. 火烧信号对种子萌发影响的研究进展[J]. 植物生态学报, 2021, 45(11): 1177-1190. |
| [12] | 张楠,刘自广,孙世臣,刘圣怡,林建辉,彭疑芳,张晓旭,杨贺,岑曦,吴娟. 拟南芥AtR8 lncRNA对盐胁迫响应及其对种子萌发的调节作用[J]. 植物学报, 2020, 55(4): 421-429. |
| [13] | 曹栋栋,陈珊宇,秦叶波,吴华平,阮关海,黄玉韬. 水杨酸调控盐胁迫下羽衣甘蓝种子萌发的机理[J]. 植物学报, 2020, 55(1): 49-61. |
| [14] | 陈唯,曾晓贤,谢楚萍,田长恩,周玉萍. 植物内源ABA水平的动态调控机制[J]. 植物学报, 2019, 54(6): 677-687. |
| [15] | 杨立文,刘双荣,林荣呈. 光信号与激素调控种子休眠和萌发研究进展[J]. 植物学报, 2019, 54(5): 569-581. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||