

植物学报 ›› 2021, Vol. 56 ›› Issue (2): 218-231.DOI: 10.11983/CBB20141 cstr: 32102.14.CBB20141
宋松泉1,2,*( ), 刘军2,*(
), 刘军2,*( ), 杨华3, 张文虎2, 张琪2, 高家东2
), 杨华3, 张文虎2, 张琪2, 高家东2
收稿日期:2020-08-03
接受日期:2021-01-05
出版日期:2021-03-01
发布日期:2021-03-17
通讯作者:
宋松泉,刘军
作者简介:liujun@gdaas.cn基金资助:
Songquan Song1,2,*( ), Jun Liu2,*(
), Jun Liu2,*( ), Hua Yang3, Wenhu Zhang2, Qi Zhang2, Jiadong Gao2
), Hua Yang3, Wenhu Zhang2, Qi Zhang2, Jiadong Gao2
Received:2020-08-03
Accepted:2021-01-05
Online:2021-03-01
Published:2021-03-17
Contact:
Songquan Song,Jun Liu
摘要: 种子萌发是子代植株建立、生长和繁育的重要阶段, 在种子植物生命周期中起重要作用。种子休眠是在发育过程中形成的, 在生理成熟期达到峰值。种子休眠与萌发的植物激素调控可能是种子植物中一种高度保守的机制。细胞分裂素(CK)是植物体内的一种重要信号分子, 调控植物生长发育的许多方面。生物活性CK的水平由其生物合成、活化、失活、再活化和降解之间的平衡所调控, 种子的发育、休眠与萌发受生物活性CK的水平和信号转导途径调控。该文综述了CK的生物合成与代谢、信号转导以及对种子发育、休眠与萌发的调控作用, 提出了在本领域需要进一步研究的科学问题, 旨在为理解CK调控种子发育、休眠与萌发的分子机理提供参考。
宋松泉, 刘军, 杨华, 张文虎, 张琪, 高家东. 细胞分裂素调控种子发育、休眠与萌发的研究进展. 植物学报, 2021, 56(2): 218-231.
Songquan Song, Jun Liu, Hua Yang, Wenhu Zhang, Qi Zhang, Jiadong Gao. Research Progress in Seed Development, Dormancy and Germination Regulated by Cytokinin. Chinese Bulletin of Botany, 2021, 56(2): 218-231.
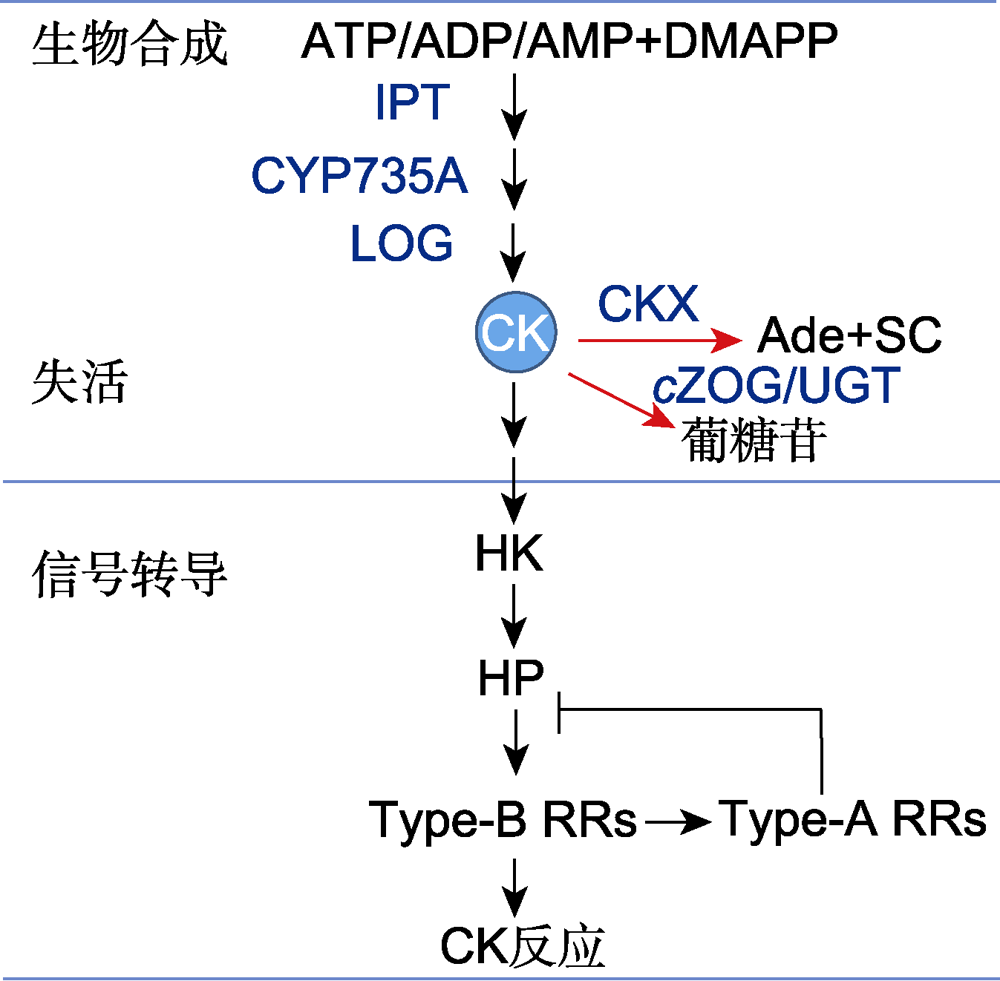
图1 细胞分裂素(CK)代谢和信号转导模型(改自Kieber and Schaller, 2018; Zubo and Schaller, 2020) Ade: 腺嘌呤; ADP: 腺苷二磷酸; AMP: 腺苷一磷酸; ATP: 腺苷三磷酸; CKX: CK氧化酶/脱氢酶; CYP735A: 细胞色素P450单加氧酶; cZOG: 顺式玉米素-O-葡糖基转移酶; DMAPP: 二甲基丙烯基二磷酸; HK: 组氨酸激酶; HP: 组氨酸磷酸转移蛋白; IPT: 异戊烯基转移酶; LOG: CK核苷5′-单磷酸磷酸核糖水解酶; SC: 侧链; Type-B RRs: 类型-B反应调控因子; Type-A RRs: 类型-A反应调控因子; UGT: UDP糖基转移酶
Figure 1 A model of cytokinin (CK) metabolism and signaling transduction (modified from Kieber and Schaller, 2018; Zubo and Schaller, 2020) Ade: Adenine; ADP: Adenosine diphosphate; AMP: Adenosine monophosphate; ATP: Adenosine triphosphate; CKX: CK oxidase/dehydrogenase; CYP735A: Cytochrome P450 monooxygenase; cZOG: cis-Zeatin-O-glucosyltransferase; DMAPP: Dimethylallyl diphosphate; HK: Histidin (His) kinase; HP: His phosphotransfer protein; IPT: Isopentenyltransferase; LOG: CK nucleoside 5′-monophosphate phosphoribohydrolase; SC: Sidechain; Type-B RRs: Type-B response regulators; Type-A RRs: Type-A response regulators; UGT: UDP glycosyltransferase
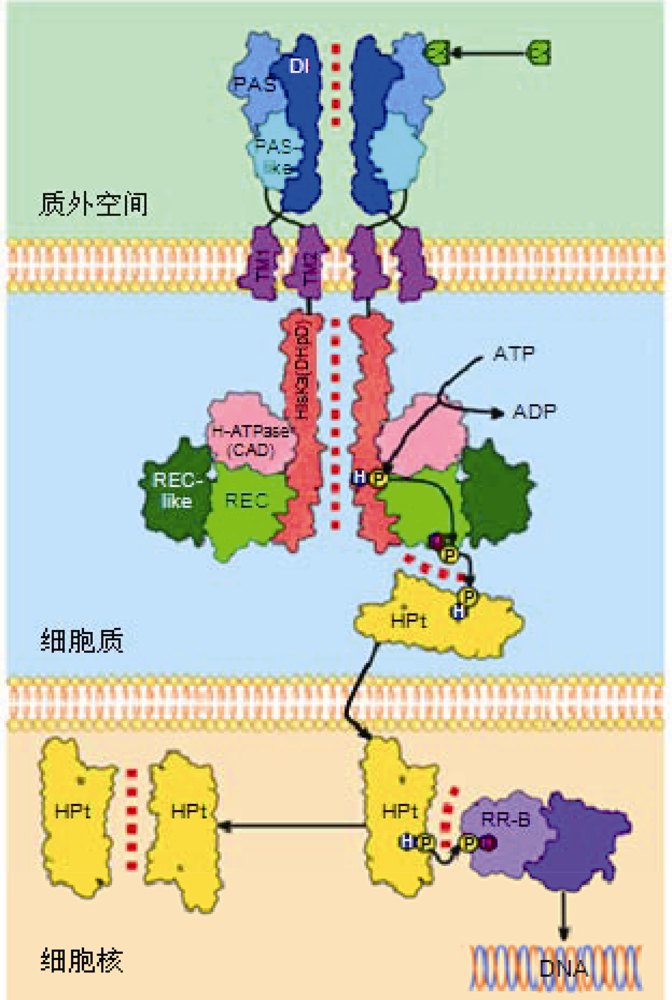
图2 细胞分裂素信号转导途径中的分子间相互作用(改自Lomin et al. 2018; Arkhipov et al. 2019) 细胞分裂素信号被膜定位的组氨酸(His)激酶受体感受, 通过His磷酸转移蛋白被传递到细胞核, 从而激活细胞核中的转录因子家族。CK: 细胞分裂素; ATP: 腺苷三磷酸; ADP: 腺苷二磷酸; P: 磷酸; D: 保守的天冬氨酸; H: 保守的His; DI: 感受器模块的二聚体界面结构域; PAS和PAS-like: 感受器模块的CHASE结构域的子结构域; TM1和TM2: 跨膜结构域1和2; HisKA (DHpD): His激酶A结构域(二聚合作用和His磷酸转移结构域); H-ATPase (CAD): 腺苷三磷酸酶结构域(催化与ATP结合结构域); REC-like: 类接收器结构域; REC: 接收器结构域; HPt: 含有His的磷酸转移蛋白(磷酸传递蛋白); RR-B: B型反应调控因子(转录因子)。蛋白-蛋白相互作用(PPI)用红色虚线表示。
Figure 2 Intermolecular interactions in the cytokinin signaling pathway (modified from Lomin et al. 2018; Arkhipov et al. 2019) Cytokinin signaling is perceived by membrane-localized Histidine (His) kinase receptors and is transduced to nucleus through a His phosphotransfer protein to activate a family of transcription factors in the nucleus. CK: Cytokinin; ATP: Adenosine triphosphate; ADP: Adenosine diphosphate; P: Phosphate; D: Conserved Aspartate; H: Conserved His; DI: Dimerization interface domain of the sensor module; PAS and PAS-like: Subdomains of the CHASE domain of the sensor module; TM1 and TM2: Transmembrane domain 1 and 2; HisKA (DHpD): His kinase A domain (dimerization and His phosphotransfer domain); H-ATPase (CAD): Adenosine triphosphatase domain (catalytic and ATP-binding domain); REC-like: Receiver-like domain; REC: Receiver domain; HPt: His-containing phosphotransfer protein (phosphotransmitter); RR-B: Type B response regulator (transcription factor). Protein-protein interactions (PPI) are indicated by a red dotted line.
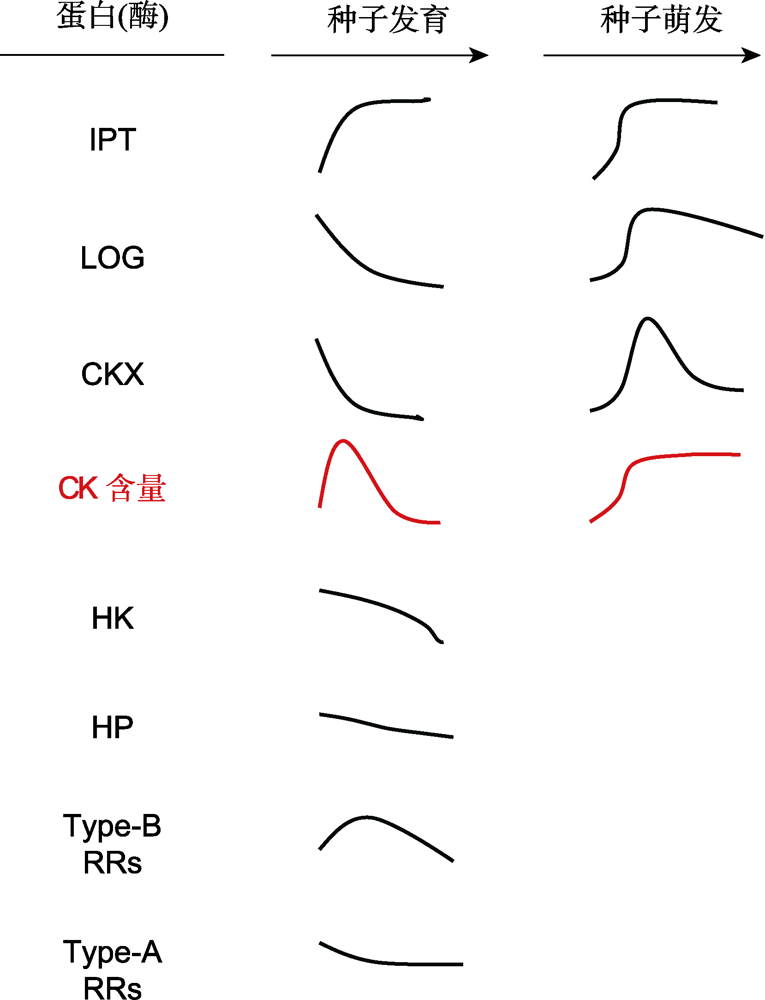
图3 种子发育和萌发过程中与细胞分裂素(CK)代谢和信号转导组分有关的蛋白(酶)的变化模式图(改自Jameson et al. 2016; Tuan et al. 2019; Nguyen et al. 2020) IPT: 异戊烯基转移酶; LOG: CK核苷5′-单磷酸磷酸核糖水解酶; CKX: CK氧化酶/脱氢酶; HK: 组氨酸激酶; HP: 组氨酸磷酸转移蛋白; Type-B RRs: 类型-B反应调控因子; Type-A RRs: 类型-A反应调控因子
Figure 3 The model of the change of proteins (enzymes) involved in cytokinin (CK) metabolism and signaling components during seed development and germination (modified from Jameson et al. 2016; Tuan et al. 2019; Nguyen et al. 2020) IPT: Isopentenyltransferase; LOG: CK nucleoside 5′-monophosphate phosphoribohydrolase; CKX: CK oxidase/ dehydrogenase; HK: Histidin (His) kinase; HP: His phosphotransfer protein; Type-B RRs: Type-B response regulators; Type-A RRs: Type-A response regulators
| [1] | 邓志军, 宋松泉 (2008). ABA对黑黄檀种子萌发的抑制作用以及其他植物激素对ABA的拮抗作用. 云南植物研究 30,440-446. |
| [2] | 邓志军, 宋松泉, 艾训儒, 姚兰 (2019). 植物种子保存和检测的原理与技术. 北京: 科学出版社. pp.22-74. |
| [3] | 宋松泉, 刘军, 黄荟, 伍贤进, 徐恒恒, 张琪, 李秀梅, 梁娟 (2020a). 赤霉素代谢与信号转导及其调控种子萌发与休眠的分子机制. 中国科学: 生命科学 50,599-615. |
| [4] | 宋松泉, 刘军, 徐恒恒, 刘旭, 黄荟 (2020b). 脱落酸代谢与信号传递及其调控种子休眠与萌发的分子机制. 中国农业科学 53,857-873. |
| [5] |
徐恒恒, 黎妮, 刘树君, 王伟青, 王伟平, 张红, 程红焱, 宋松泉 (2014). 种子萌发及其调控的研究进展. 作物学报 40,1141-1156.
DOI URL |
| [6] |
Araújo S, Pagano A, Dondi D, Lazzaroni S, Pinela E, Macovei A, Balestrazzi A (2019). Metabolic signatures of germination triggered by kinetin in Medicago truncatula. Sci Rep 9, 10466.
DOI URL |
| [7] |
Arkhipov DV, Lomin SN, Myakushina YA, Savelieva EM, Osolodkin DI, Romanov GA (2019). Modeling of protein-protein interactions in cytokinin signal transduction. Int J Mol Sci 20,2096.
DOI URL |
| [8] | Baskin CC, Baskin JM (2014). Seeds:Ecology, Biogeography, and Evolution of Dormancy and Germination, 2nd edn. Amsterdam: Academic Press. pp.5-77. |
| [9] |
Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110,E435- E444.
DOI URL |
| [10] | Bewley JD, Bradford KJ, Hilhorst HWM, Nonogaki H (2013). Physiology of Development,Germination and Dormancy, 3rd edn. New York: Springer. pp.27-83. |
| [11] |
Cairns JRK, Esen A (2010). β-glucosidases. Cell Mol Life Sci 67,3389-3405.
DOI URL |
| [12] |
Chen L, Zhao JQ, Song JC, Jameson PE (2020). Cytokinin dehydrogenase: a genetic target for yield improvement in wheat. Plant Biotechnol J 18,614-630.
DOI PMID |
| [13] |
Chitnis VR, Gao F, Yao Z, Jordan MC, Park S, Ayele BT (2014). After-ripening induced transcriptional changes of hormonal genes in wheat seeds: the cases of brassinosteroids, ethylene, cytokinin and salicylic acid. PLoS One 9,e87543.
DOI URL |
| [14] | Corbineau F, Xia Q, Bailly C, EI-Maarouf-Bouteau H (2014). Ethylene, a key factor in the regulation of seed dormancy. Front Plant Sci 5,539. |
| [15] |
Day RC, Herridge RP, Ambrose BA, Macknight RC (2008). Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol 148,1964-1984.
DOI URL |
| [16] |
Deng Y, Dong HL, Mu JY, Ren B, Zheng BL, Ji ZD, Yang WC, Liang Y, Zuo JR (2010). Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22,1232-1248.
DOI URL |
| [17] | Eastwood D, Tavener RJA, Laidman DL (1969). Sequential action of cytokinin and gibberellic acid in wheat aleurone tissue. Nature 221,1267. |
| [18] |
Feng J, Wang C, Chen QG, Chen H, Ren B, Li XM, Zuo JR (2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat Commun 4,1529.
DOI URL |
| [19] |
Frébort I, Kowalska M, Hluska T, Frébortová J, Galuszka P (2011). Evolution of cytokinin biosynthesis and degradation. J Exp Bot 62,2431-2452.
DOI URL |
| [20] |
Guan CM, Wang XC, Feng J, Hong SL, Liang Y, Ren B, Zuo JR (2014). Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of ABSCISIC ACID INSENSITIVE 5 protein in Arabidopsis. Plant Physiol 164, 1515-1526.
DOI URL |
| [21] |
Hallmark HT, Rashotte AM (2019). Review—cytokinin response factors: responding to more than cytokinin. Plant Sci 289,110251.
DOI URL |
| [22] |
Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H (2008). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59,75-83.
DOI URL |
| [23] |
Hošek P, Hoyerová K, Kiran NS, Dobrev PI, Zahajská L, Filepová R, Motyka V, Müller K, Kamínek M (2020). Distinct metabolism of N-glucosides of isopentenyladenine and trans-zeatin determines cytokinin metabolic spectrum in Arabidopsis. New Phytol 225,2423-2438.
DOI URL |
| [24] |
Hothorn M, Dabi T, Chory J (2011). Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat Chem Biol 7,766-768.
DOI URL |
| [25] |
Hou BK, Lim EK, Higgins GS, Bowles DJ (2004). N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279,47822-47832.
DOI URL |
| [26] |
Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18,3073-3087.
PMID |
| [27] | Hwang I, Sheen J, Müller B (2012). Cytokinin signaling networks. Annu Rev Plant Biol 63,353-380. |
| [28] |
Jameson PE, Dhandapani P, Novak O, Song JC (2016). Cytokinins and expression of SWEET, SUT, CWINV and AAP genes increase as pea seeds germinate. Int J Mol Sci 17,2013.
DOI URL |
| [29] |
Jameson PE, Song JC (2016). Cytokinin: a key driver of seed yield. J Exp Bot 67,593-606.
DOI URL |
| [30] |
Kakimoto T (2001). Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol 42,677-685.
PMID |
| [31] |
Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, Yamaguchi S, Sakakibara H (2004). Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J Biol Chem 279,14049-14054.
DOI URL |
| [32] |
Keshishian EA, Rashotte AM (2015). Plant cytokinin signaling. Essays Biochem 58,13-27.
DOI URL |
| [33] |
Khan AA (1968). Inhibition of gibberellic acid-induced germination by abscisic acid and reversal by cytokinins. Plant Physiol 43,1463-1465.
PMID |
| [34] |
Kieber JJ, Schaller GE (2014). Cytokinins. Arabidopsis Book 12,e0168.
DOI URL |
| [35] |
Kieber JJ, Schaller GE (2018). Cytokinin signaling in plant development. Development 145,dev149344.
DOI URL |
| [36] |
Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445,652-655.
DOI URL |
| [37] |
Li YJ, Cheng HY, Song SQ (2009). Effects of temperature, after-ripening, stratification, and scarification plus hormone treatments on dormancy release and germination of Acer truncatum seeds. Seed Sci Technol 37,554-562.
DOI URL |
| [38] |
Liu HX, Stone SL (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22,2630-2641.
DOI URL |
| [39] |
Liu XD, Zhang H, Zhao Y, Feng ZY, Li Q, Yang HQ, Luan S, Li JM, He ZH (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110,15485-15490.
DOI URL |
| [40] |
Liu ZN, Yuan L, Song XY, Yu XL, Sundaresan V (2017). AHP2, AHP3, and AHP5 act downstream of CKI1 in Arabidopsis female gametophyte development. J Exp Bot 68,3365-3373.
DOI URL |
| [41] |
Lomin SN, Krivosheev DM, Steklov MY, Arkhipov DV, Osolodkin DI, Schmülling T, Romanov GA (2015). Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J Exp Bot 66,1851-1863.
DOI URL |
| [42] |
Lomin SN, Myakushina YA, Kolachevskaya OO, Getman IA, Arkhipov DV, Savelieva EM, Osolodkin DI, Romanov GA (2018). Cytokinin perception in potato: new features of canonical players. J Exp Bot 69,3839-3853.
DOI URL |
| [43] |
Lur HS, Setter TL (1993). Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin). Plant Physiol 103,273- 280.
DOI URL |
| [44] |
Mähönen AP, Higuchi M, Tormakangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T (2006). Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol 16, 1116-1122.
PMID |
| [45] |
Marín-de la Rosa N, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Gronlund AL, Wanchoo- Kohli A, Thomas SG, Bennett MJ, Lohmann JU, Blázquez MA, Alabadí D (2015). Genome wide binding site analysis reveals transcriptional coactivation of cytokinin- responsive genes by DELLA proteins. PLoS Genet 11,e1005337.
DOI URL |
| [46] | Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37,128-138. |
| [47] | Mok MC, Martin RC, Dobrev PI, Vanková R, Ho PS, Yonekura-Sakakibara K, Sakakibara H, Mok DWS (2005). Topolins and hydroxylated thidiazuron derivatives are substrates of cytokinin O-glucosyltransferase with position specificity related to receptor recognition. Plant Physiol 137,1057-1066. |
| [48] | Müller B, Sheen J (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453,1094-1097. |
| [49] | Nguyen HN, Perry L, Kisiala A, Olechowski H, Emery RJN (2020). Cytokinin activity during early kernel development corresponds positively with yield potential and later stage ABA accumulation in field-grown wheat ( Triticum aestivum L.). Planta 252,76. |
| [50] | Nonogaki H (2014). Seed dormancy and germination- emerging mechanisms and new hypotheses. Front Plant Sci 5,233. |
| [51] | Nonogaki H (2017). Seed biology updates—highlights and new discoveries in seed dormancy and germination research. Front Plant Sci 8,524. |
| [52] | Nonogaki H (2019). Seed germination and dormancy: the classic story, new puzzles, and evolution. J Integr Plant Biol 61,541-563. |
| [53] | Pekárová B, Szmitkowska A, Dopitová R, Degtjarik O, Žídek L, Hejátko J (2016). Structural aspects of multistep phosphorelay-mediated signaling in plants. Mol Plant 9,71-85. |
| [54] | Pekarova B, Szmitkowska A, Houser J, Wimmerova M, Hejátko J (2018). Cytokinin and ethylene signaling. In: Hejátko J, Hakoshima T, eds. Plant Structural Biology:Hormonal Regulations. Cham: Springer. pp.165-200. |
| [55] | Ren B, Liang Y, Deng Y, Chen QG, Zhang J, Yang XH, Zuo JR (2009). Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res 19, 1178- 1190. |
| [56] | Rijavec T, Dermastia M (2010). Cytokinins and their function in developing seeds. Acta Chim Slov 57,617-629. |
| [57] | Romanov GA, Lomin SN, Schmülling T (2018). Cytokinin signaling: from the ER or from the PM? That is the question! New Phytol 218,41-53. |
| [58] | Sakakibara H (2006). Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57,431-449. |
| [59] | Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, Asami T, Okada K, Kamiya Y, Yamaya T, Yamaguchi S (2005). Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc Natl Acad Sci USA 102,9972-9977. |
| [60] | Sakano Y, Okada Y, Matsunaga A, Suwama T, Kaneko T, Ito K, Noguchi H, Abe I (2004). Molecular cloning, expression, and characterization of adenylate isopentenyltransferase from hop ( Humulus lupulus L.). Phytochemistry 65,2439-2446. |
| [61] | Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP, Kurata N, Mizuno T, Pareek A, Shiu SH, Wu P, Yip WK (2007). Nomenclature for two-component signaling elements of rice. Plant Physiol 143,555-557. |
| [62] | Shu K, Liu XD, Xie Q, He ZH (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9,34-45. |
| [63] | Shuai HW, Meng YJ, Luo XF, Chen F, Zhou WG, Dai YJ, Qi Y, Du JB, Yang F, Liu J, Yang WY, Shu K (2017). Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Sci Rep 7,12620. |
| [64] | Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov GA, Strnad M, Schmülling T (2004). Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol 45,1299-1305. |
| [65] | Steklov MY, Lomin SN, Osolodkin DI, Romanov GA (2013). Structural basis for cytokinin receptor signaling: an evolutionary approach. Plant Cell Rep 32,781-793. |
| [66] | Strnad M (1997). The aromatic cytokinins. Physiol Plant 101,674-688. |
| [67] | Takei K, Sakakibara H, Sugiyama T (2001). Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276,26405-26410. |
| [68] | Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A (2007). ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48,263-277. |
| [69] | Tarkowska D, Doležal K, Tarkowski P, Åstot C, Holub J, Fuksová K, Schmülling T, Sandberg G, Strnad M (2003). Identification of new aromatic cytokinins in Arabidopsis thaliana and Populus × canadensis leaves by LC- (+) ESI-MS and capillary liquid chromatography/frit- fast atom bombardment mass spectrometry. Physiol Plant 117,579-590. |
| [70] | To JPC, Deruère J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2007). Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19,3901-3914. |
| [71] | Tuan PA, Yamasaki Y, Kanno Y, Seo M, Ayele BT (2019). Transcriptomics of cytokinin and auxin metabolism and signaling genes during seed maturation in dormant and non-dormant wheat genotypes. Sci Rep 9,3983. |
| [72] | Wang YP, Li L, Ye TT, Zhao SJ, Liu Z, Feng YQ, Wu Y (2011). Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J 68,249-261. |
| [73] | Wulfetange K, Lomin SN, Romanov GA, Stolz A, Heyl A, Schmülling T (2011). The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol 156,1808-1818. |
| [74] | Wybouw B, De Rybel B (2019). Cytokinin—a developing story. Trends Plant Sci 24,177-185. |
| [75] | Zalabák D, Galuszka P, Mrízová K, Podlešáková K, Gu RL, Frébortová J (2014). Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant Physiol Biochem 74,283-293. |
| [76] | Zdarska M, Dobisová T, Gelová Z, Pernisová M, Dabravolski S, Hejátko J (2015). Illuminating light, cytokinin, and ethylene signaling crosstalk in plant development. J Exp Bot 66,4913-4931. |
| [77] | Zschiedrich CP, Keidel V, Szurmant H (2016). Molecular mechanisms of two-component signal transduction. J Mol Biol 428,3752-3775. |
| [78] | Zubko E, Adams CJ, Macháèková I, Malbeck J, Scollan C, Meyer P (2002). Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J 29,797-808. |
| [79] | Zubo YO, Blakley IC, Yamburenko MV, Worthen JM, Street IH, Franco-Zorrilla JM, Zhang WJ, Hill K, Raines T, Solano R, Kieber JJ, Loraine AE, Schaller GE (2017). Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc Natl Acad Sci USA 114,E5995-E6004. |
| [80] | Zubo YO, Schaller GE (2020). Role of the cytokinin-activated type-B response regulators in hormone crosstalk. Plants 9,166. |
| [1] | 平晓燕, 杜毅倩, 赖仕蓉, 孔梦桥, 余国杰. 植物应对食草动物采食的化学防御策略研究进展[J]. 植物生态学报, 2025, 49(5): 667-680. |
| [2] | 马亮, 杨永青, 郭岩. “后绿色革命”基因——助力培育“气候智能”作物新品种[J]. 植物学报, 2025, 60(4): 489-498. |
| [3] | 褚晓琳, 张全国. 演化速率假说的实验验证研究进展[J]. 生物多样性, 2025, 33(4): 25019-. |
| [4] | 叶灿, 姚林波, 金莹, 高蓉, 谭琪, 李旭映, 张艳军, 陈析丰, 马伯军, 章薇, 张可伟. 水稻水杨酸代谢突变体高通量筛选方法的建立与应用[J]. 植物学报, 2025, 60(4): 1-0. |
| [5] | 莫笑梅, 张琪, 杨嘉欣, 郑国, 胡中民, 张晓珂, 梁思维, 崔淑艳. 北方典型草地土壤线虫代谢速率及能量流动对氮沉降和降水模式改变的响应[J]. 生物多样性, 2025, 33(3): 24341-. |
| [6] | 陈楠, 张全国. 实验进化研究途径[J]. 生物多样性, 2024, 32(9): 24171-. |
| [7] | 周玉滢, 陈辉, 刘斯穆. 植物非典型Aux/IAA蛋白应答生长素研究进展[J]. 植物学报, 2024, 59(4): 651-658. |
| [8] | 何璐梅, 马伯军, 陈析丰. 植物执行者抗病基因研究进展[J]. 植物学报, 2024, 59(4): 671-680. |
| [9] | 孙苗苗, 张蔚, 张林霞, 霍竣涛, 李志能, 刘国锋. 矮牵牛花朵大小遗传规律及相关基因的表达分析[J]. 植物学报, 2024, 59(3): 422-432. |
| [10] | 刘笑, 杜琬莹, 张云秀, 唐成名, 李华伟, 夏海勇, 樊守金, 孔令安. NO3-缓解小麦根部NH4+毒性机理(长英文摘要)[J]. 植物学报, 2024, 59(3): 397-413. |
| [11] | 车佳航, 李纬楠, 秦英之, 陈金焕. 木本植物叶色变异机制研究进展[J]. 植物学报, 2024, 59(2): 319-328. |
| [12] | 赵常提, 夏青霖, 田地, 陈冰瑞, 朱瑞德, 刘宵含, 俞果, 吉成均. 长期氮添加对温带落叶阔叶林优势植物叶片次生代谢产物的影响[J]. 植物生态学报, 2024, 48(12): 1576-1588. |
| [13] | 商慧颖, 翟云雁, 鬲晓敏, 刘帅, 王尚林, 周涛, 柏国清. 基于广泛靶向代谢组学揭示不同时期漆树生漆中代谢物变化规律[J]. 植物学报, 2024, 59(1): 66-74. |
| [14] | 朱宝, 赵江哲, 张可伟, 黄鹏. 水稻细胞分裂素氧化酶9参与调控水稻叶夹角发育[J]. 植物学报, 2024, 59(1): 10-21. |
| [15] | 朱璐, 袁冲, 刘义飞. 植物次生代谢产物生物合成基因簇研究进展[J]. 植物学报, 2024, 59(1): 134-143. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||