

植物学报 ›› 2024, Vol. 59 ›› Issue (3): 422-432.DOI: 10.11983/CBB23141 cstr: 32102.14.CBB23141
孙苗苗1, 张蔚1,2, 张林霞1, 霍竣涛1,2, 李志能2,*( ), 刘国锋1,*(
), 刘国锋1,*( )
)
收稿日期:2023-10-16
接受日期:2024-03-18
出版日期:2024-05-10
发布日期:2024-05-10
通讯作者:
E-mail: 基金资助:
Miaomiao Sun1, Wei Zhang1,2, Linxia Zhang1, Juntao Huo1,2, Zhineng Li2,*( ), Guofeng Liu1,*(
), Guofeng Liu1,*( )
)
Received:2023-10-16
Accepted:2024-03-18
Online:2024-05-10
Published:2024-05-10
Contact:
E-mail: 摘要: 花朵大小是植物进化及物种形成过程中的关键因子, 也是决定植物观赏价值的重要性状, 研究其遗传规律与调控机制具有重要的科学意义和实用价值。以不同花朵大小的矮牵牛(Petunia hybrida)自交系和原生种为材料, 配制大花×中花和大花×小花杂交组合, 构建遗传群体, 研究其花朵大小的遗传规律。结果表明, 大花自交系W与腋花矮牵牛S26 (中花)杂交F1群体均为大花, F2群体中大花与中花的分离比接近3:1, BC1回交群体中大花与中花的分离比接近1:1; 大花自交系W与中花自交系S杂交F1群体均为大花, F2群体花朵大小出现分离, 大花与中花的比例接近2:1; 大花自交系W与膨大矮牵牛S6 (小花)杂交F1群体均为中花, F2群体则出现花径从大到小的连续分布。利用主基因+多基因混合模型分析, 按照AIC值最小的标准选出W × S26组合的最优模型为1MG-AD, W × S的最优模型为2MG-EAD, 由此判定W大花对S26中花由1对主显性基因控制, 具加性-显性效应, W大花对S中花则由2对主基因控制, 具等加性-显性效应。此外, 根据前期大、小花转录组分析结果, 选取9个可能调控花朵大小的基因, 通过qPCR检测了它们在不同花朵大小的矮牵牛株系花瓣中的表达水平, 结果发现细胞分裂素受体基因PhHK以及响应细胞分裂素信号的type-A RRs基因在大花中的表达水平普遍高于中花和小花, 表明细胞分裂素信号途径可能是参与调控矮牵牛大花性状的关键因素。
孙苗苗, 张蔚, 张林霞, 霍竣涛, 李志能, 刘国锋. 矮牵牛花朵大小遗传规律及相关基因的表达分析. 植物学报, 2024, 59(3): 422-432.
Miaomiao Sun, Wei Zhang, Linxia Zhang, Juntao Huo, Zhineng Li, Guofeng Liu. Inheritance Analysis of Flower Size and Expression of Related Genes in Petunia hybrida. Chinese Bulletin of Botany, 2024, 59(3): 422-432.

图1 不同花朵大小矮牵牛实验材料 W、DHZ、KZ、W115、S、SW、HC和SP: 矮牵牛自交系; S26: 腋花矮牵牛; S6: 膨大矮牵牛。Bar=1 cm
Figure 1 The tested materials of petunia with different flower sizes W, DHZ, KZ, W115, S, SW, HC, and SP: The inbred lines of Petunia hybrida; S26: P. axillaris; S6: P. inflata. Bar=1 cm
| Gene name | Primer name | Primer sequence (5'−3') |
|---|---|---|
| PhARR496 | qPhARR496-F | GCAACAACAAAGAAGCAGAGCAGTT |
| qPhARR496-R | GGTGATGATGGTAATGGCTGTGTCA | |
| PhARR828 | qPhARR828-F | CATCTTCCTCGCTCTCGCTCTC |
| qPhARR828-R | TGAGTCGTCGTGTTGGTGAATGT | |
| PhARR970 | qPhARR970-F | ACTCCTCTTCCTCATCAGTCATCATCA |
| qPhARR970-R | GCTGCTGCCATTCCCATTGTTG | |
| PhARR1029 | qPhARR1029-F | GCAAGAGCATGAACAGAACAAGTCA |
| qPhARR1029-R | CCAGGCATACAGTAGTCCGTGATAA | |
| PhARGOS057 | qPhARGOS057-F | TCTCTTCCATCATACGCTTTGGTCAA |
| qPhARGOS057-R | AGGCAAGAACCATAGGAGATTGTTGA | |
| PhARGOS459 | qPhARGOS459-F | GCACATGGAATCATCAGCAGAAGC |
| qPhARGOS459-R | GGTGGCAATGGTGGAAGGATCAA | |
| PhHK | qPhHK-F | CCAAGTTCTTTGAATCCAAGCCTCAC |
| qPhHK-R | AATAAGAATGCAGCACCAGCAGAATG | |
| PhTCP3b | qPhTCP3b-F | CCTCTCTGCTCACACAGCCATTC |
| qPTCP3b-R | GCCCAGCAATACTGTCAACTAAACT | |
| PhTCP4a | qPhTCP4a-F | TGACCAAGCAGCCCTTTTCTCT |
| qPhTCP4a-R | TGGAACTCTGTGGTGAAATGTCTGT |
表1 实时荧光定量PCR引物
Table 1 Primers used for quantitative real-time PCR
| Gene name | Primer name | Primer sequence (5'−3') |
|---|---|---|
| PhARR496 | qPhARR496-F | GCAACAACAAAGAAGCAGAGCAGTT |
| qPhARR496-R | GGTGATGATGGTAATGGCTGTGTCA | |
| PhARR828 | qPhARR828-F | CATCTTCCTCGCTCTCGCTCTC |
| qPhARR828-R | TGAGTCGTCGTGTTGGTGAATGT | |
| PhARR970 | qPhARR970-F | ACTCCTCTTCCTCATCAGTCATCATCA |
| qPhARR970-R | GCTGCTGCCATTCCCATTGTTG | |
| PhARR1029 | qPhARR1029-F | GCAAGAGCATGAACAGAACAAGTCA |
| qPhARR1029-R | CCAGGCATACAGTAGTCCGTGATAA | |
| PhARGOS057 | qPhARGOS057-F | TCTCTTCCATCATACGCTTTGGTCAA |
| qPhARGOS057-R | AGGCAAGAACCATAGGAGATTGTTGA | |
| PhARGOS459 | qPhARGOS459-F | GCACATGGAATCATCAGCAGAAGC |
| qPhARGOS459-R | GGTGGCAATGGTGGAAGGATCAA | |
| PhHK | qPhHK-F | CCAAGTTCTTTGAATCCAAGCCTCAC |
| qPhHK-R | AATAAGAATGCAGCACCAGCAGAATG | |
| PhTCP3b | qPhTCP3b-F | CCTCTCTGCTCACACAGCCATTC |
| qPTCP3b-R | GCCCAGCAATACTGTCAACTAAACT | |
| PhTCP4a | qPhTCP4a-F | TGACCAAGCAGCCCTTTTCTCT |
| qPhTCP4a-R | TGGAACTCTGTGGTGAAATGTCTGT |
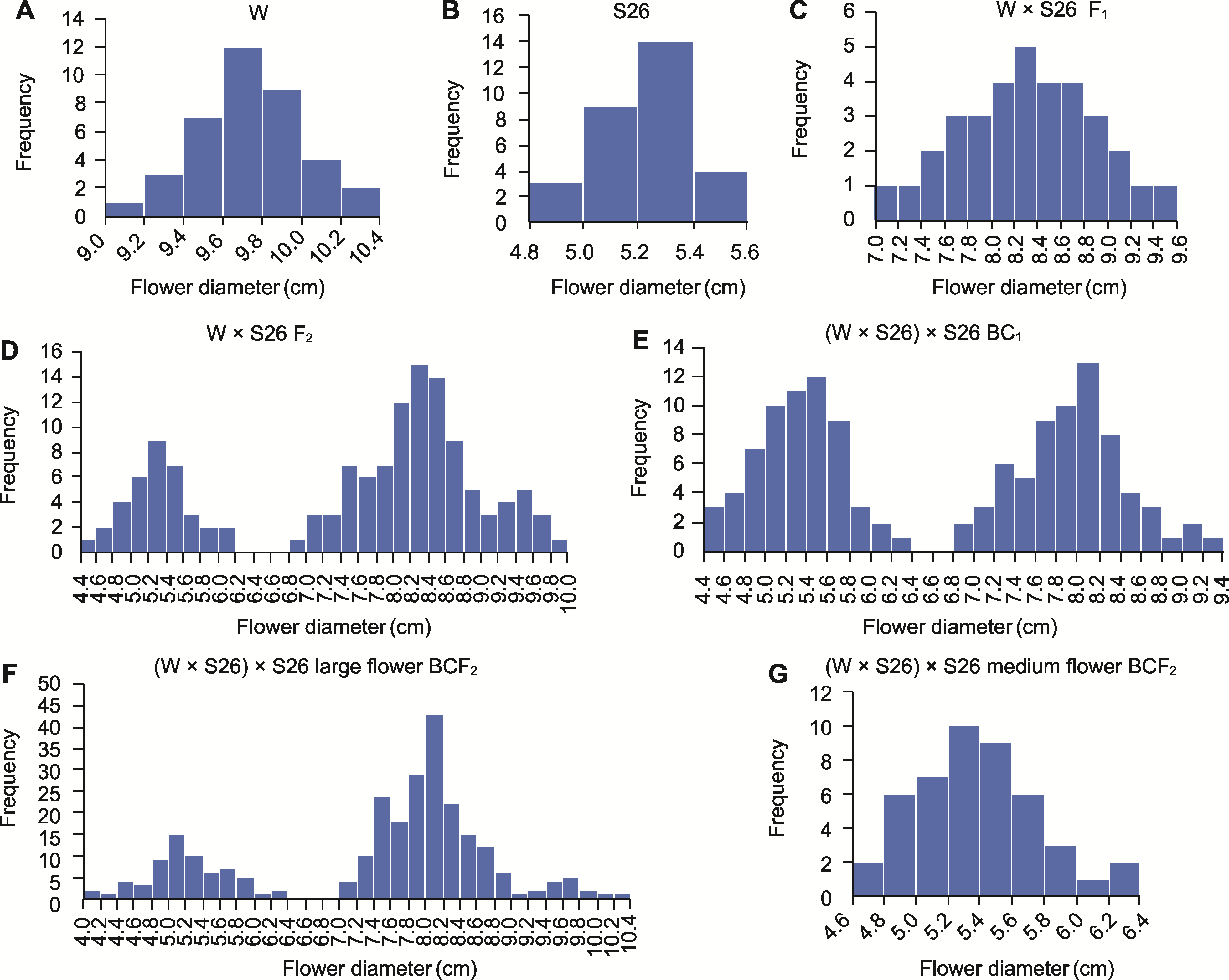
图2 W × S26亲本及不同后代群体花径的频数分布 (A) 母本W; (B) 父本S26; (C) F1群体; (D) F2群体; (E) BC1群体; (F) 大花BCF2群体; (G) 中花BCF2群体
Figure 2 Frequency distribution histogram of flower diameter for parents and filial generations of W × S26 (A) Female parent W; (B) Male parent S26; (C) F1 generation; (D) F2 generation; (E) BC1 generation; (F) BCF2 generation from large flower; (G) BCF2 generation from medium flower
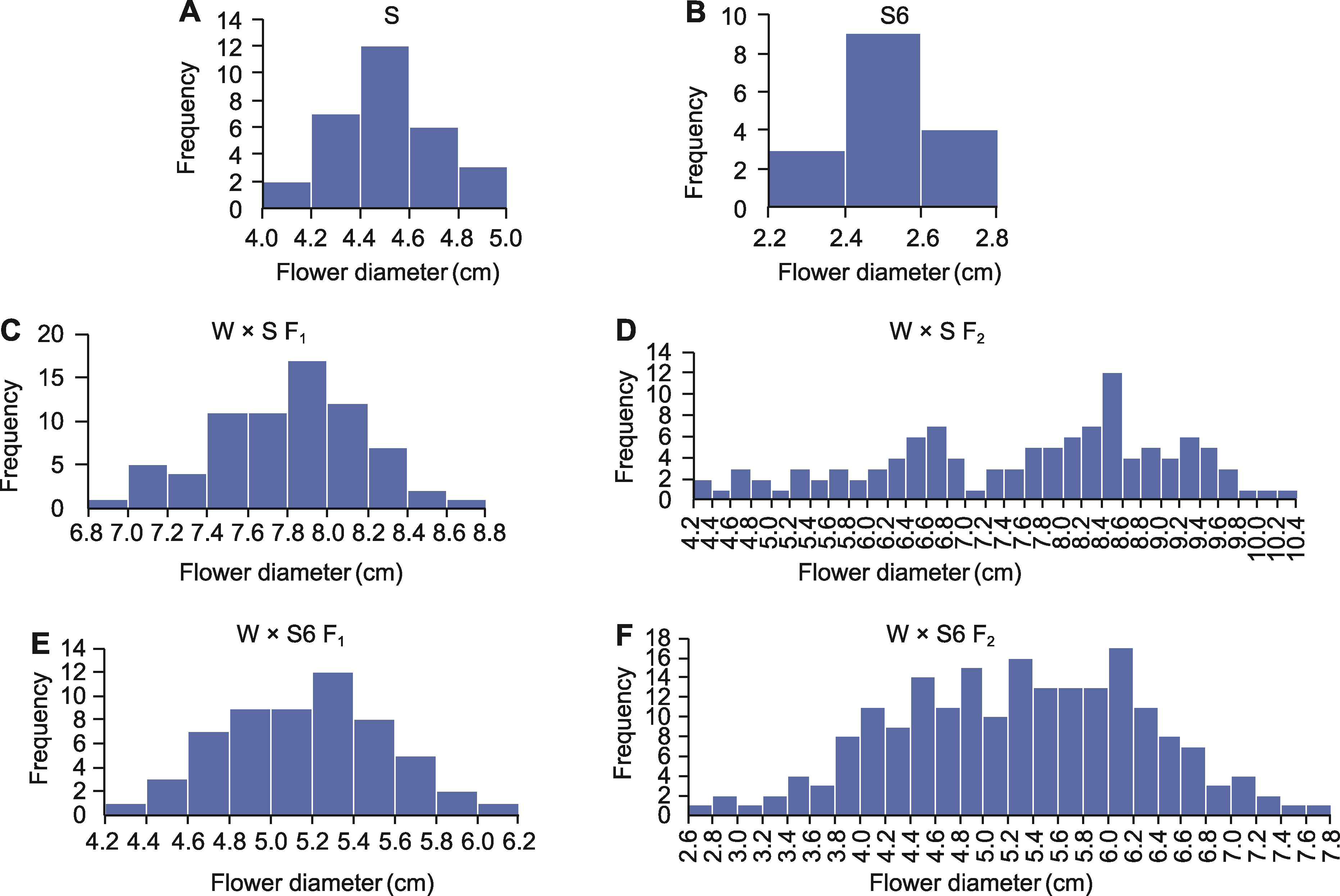
图3 W × S和W × S6亲本及F1、F2群体花径频数的分布 (A) 亲本S; (B) 亲本S6; (C) W × S F1群体; (D) W × S F2群体; (E) W × S6 F1群体; (F) W × S6 F2群体
Figure 3 Frequency distribution histogram of flower diameter for parent, F1, and F2 generations of W × S and W × S6 (A) Parent S; (B) Parent S6; (C) F1 generation of W × S; (D) F2 generation of W × S; (E) F1 generation of W × S6; (F) F2 generation of W × S6
| Cross combination | Genetic model | Maximum likelihood value | AIC | Number of significant levels (P<0.05) | ||||
|---|---|---|---|---|---|---|---|---|
| U12 | U22 | U32 | nW2 | Dn | ||||
| W × S26 | 1MG-AD | -599.9208 | 1207.8420 | 0 | 0 | 0 | 0 | 0 |
| 2MG-ADI | -596.0287 | 1212.0570 | 0 | 0 | 0 | 0 | 0 | |
| MX1-AD-AD | -596.8717 | 1211.7440 | 0 | 0 | 0 | 0 | 0 | |
| W × S | 2MG-EAD | -242.0814 | 492.1628 | 0 | 0 | 0 | 0 | 0 |
| MX2-EAD-AD | -245.5385 | 495.0770 | 0 | 0 | 0 | 0 | 0 | |
| 2MG-EA | -244.9837 | 497.9675 | 0 | 0 | 0 | 0 | 0 | |
表2 各遗传模型极大似然值(MLV)、AIC (Akaike information criterion)值和达到显著水平个数
Table 2 Maximum likelihood value (MLV), Akaike information criterion (AIC) value, and the number of reaching significant levels for each genetic model
| Cross combination | Genetic model | Maximum likelihood value | AIC | Number of significant levels (P<0.05) | ||||
|---|---|---|---|---|---|---|---|---|
| U12 | U22 | U32 | nW2 | Dn | ||||
| W × S26 | 1MG-AD | -599.9208 | 1207.8420 | 0 | 0 | 0 | 0 | 0 |
| 2MG-ADI | -596.0287 | 1212.0570 | 0 | 0 | 0 | 0 | 0 | |
| MX1-AD-AD | -596.8717 | 1211.7440 | 0 | 0 | 0 | 0 | 0 | |
| W × S | 2MG-EAD | -242.0814 | 492.1628 | 0 | 0 | 0 | 0 | 0 |
| MX2-EAD-AD | -245.5385 | 495.0770 | 0 | 0 | 0 | 0 | 0 | |
| 2MG-EA | -244.9837 | 497.9675 | 0 | 0 | 0 | 0 | 0 | |
| Cross combination (female × male) | Population types | Plant numbers | Large flower | Medium flower | Small flower | Theoretical ratio | Actual ratio | (χc²) Chi-square test | P |
|---|---|---|---|---|---|---|---|---|---|
| W × S26 | F1 | 32 | 32 | 0 | 0 | - | - | - | - |
| F2 | 134 | 98 | 36 | 0 | 3:1 | 3:1 | 0.249 | 0.618 | |
| BC1 | 129 | 67 | 62 | 0 | 1:1 | 1:1 | 0.194 | 0.660 | |
| Large flower BCF2 | 259 | 194 | 65 | 0 | 3:1 | 3:1 | 0.001 | 0.971 | |
| Medium flower BCF2 | 44 | 0 | 44 | 0 | - | - | - | - | |
| W × S | F1 | 72 | 72 | 0 | 0 | - | - | - | - |
| F2 | 115 | 76 | 39 | 0 | - | - | - | - | |
| W × S6 | F1 | 57 | 0 | 57 | 0 | - | - | - | - |
| F2 | 200 | 11 | 168 | 21 | - | - | - | - |
表3 矮牵牛杂交后代群体花朵大小分离卡方检验
Table 3 Chi square test for flower size segregation in the hybrid offsprings of petunia
| Cross combination (female × male) | Population types | Plant numbers | Large flower | Medium flower | Small flower | Theoretical ratio | Actual ratio | (χc²) Chi-square test | P |
|---|---|---|---|---|---|---|---|---|---|
| W × S26 | F1 | 32 | 32 | 0 | 0 | - | - | - | - |
| F2 | 134 | 98 | 36 | 0 | 3:1 | 3:1 | 0.249 | 0.618 | |
| BC1 | 129 | 67 | 62 | 0 | 1:1 | 1:1 | 0.194 | 0.660 | |
| Large flower BCF2 | 259 | 194 | 65 | 0 | 3:1 | 3:1 | 0.001 | 0.971 | |
| Medium flower BCF2 | 44 | 0 | 44 | 0 | - | - | - | - | |
| W × S | F1 | 72 | 72 | 0 | 0 | - | - | - | - |
| F2 | 115 | 76 | 39 | 0 | - | - | - | - | |
| W × S6 | F1 | 57 | 0 | 57 | 0 | - | - | - | - |
| F2 | 200 | 11 | 168 | 21 | - | - | - | - |
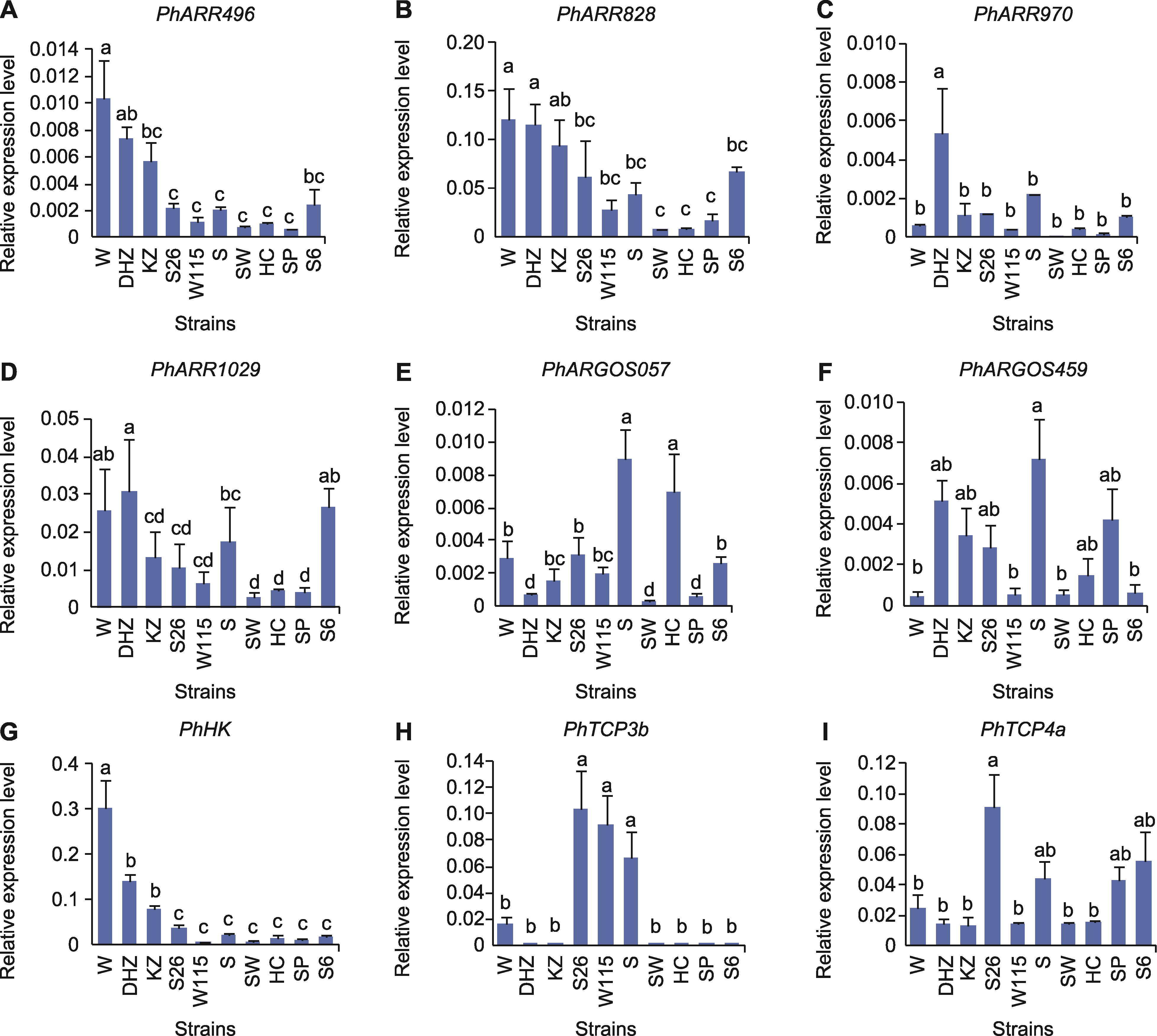
图5 参与调控花朵大小的相关基因在不同矮牵牛株系中的表达水平 误差线代表3个生物学重复的标准误差。不同小写字母表示不同株系间差异显著。
Figure 5 Expression levels of flower size regulating related genes in different strains of petunia Vertical bars represent SE of three biological replicates. Different lowercase letters indicate significant differences among strains.
| [1] |
Bartrina I, Jensen H, Novák O, Strna M, Werner T, Schmülling T (2017). Gain-of-function mutants of the cytokinin receptors AHK2 and AHK3 regulate plant organ size, flowering time and plant longevity. Plant Physiol 173, 1783-1797.
DOI PMID |
| [2] | Bombarely A, Moser M, Amrad A, Bao MZ, Bapaume L, Barry CS, Bliek M, Boersma MR, Borghi L, Bruggmann R, Bucher M, D'Agostino N, Davies K, Druege U, Dudareva N, Egea-Cortines M, Delledonne M, Fernandez-Pozo N, Franken P, Grandont L, Heslop-Harrison JS, Hintzsche J, Johns M, Koes R, Lv XD, Lyons E, Malla D, Martinoia E, Mattson NS, Morel P, Mueller LA, Muhlemann J, Nouri E, Passeri V, Pezzotti M, Qi QZ, Reinhardt D, Rich M, Richert-Pöggeler KR, Robbins TP, Schatz MC, Schranz ME, Schuurink RC, Sch- warzacher T, Spelt K, Tang HB, Urbanus SL, Vandenbussche M, Vijverberg K, Villarino GH, Warner RM, Weiss J, Yue Z, Zethof J, Quattrocchio F, Sims TL, Kuhlemeier C (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat Plants 2, 16074. |
| [3] | Cao Z, Guo YF, Yang Q, He YH, Fetouh MI, Warner RM, Deng ZA (2018). Genome-wide search for quantitative trait loci controlling important plant and flower traits in Petunia using an interspecific recombinant inbred population of Petunia axillaris and Petunia exserta. G3 8, 2309-2317. |
| [4] | Cao Z, Guo YF, Yang Q, He YH, Fetouh MI, Warner RM, Deng ZA (2019). Genome-wide identification of quantitative trait loci for important plant and flower traits in Petunia using a high-density linkage map and an interspecific recombinant inbred population derived from Petunia integrifolia and P. axillaris. Horti Res 6, 27. |
| [5] |
Crawford BCW, Nath U, Carpenter R, Coen ES (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 135, 244-253.
PMID |
| [6] | Dai SP, Bao MZ (2004). Advances in genetics and breeding of Petunia hybrida Vilm. Chin Bull Bot 21, 385-391. (in Chinese) |
| 代色平, 包满珠 (2004). 矮牵牛育种研究进展. 植物学通报 21, 385-391. | |
| [7] | Ewart L (1984). Plant breeding. In: Sink KC, ed. Petunia. Berlin: Springer. pp. 180-202. |
| [8] | Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD (2004). Pollination syndromes and floral specialization. Annu Rev Ecol Evol Syst 35, 375-403. |
| [9] |
Galliot C, Hoballah ME, Kuhlemeier C, Stuurman J (2006). Genetics of flower size and nectar volume in Petunia pollination syndromes. Planta 225, 203-212.
PMID |
| [10] | Hu YX, Poh HM, Chua NH (2006). The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J 47, 1-9. |
| [11] | Hu YX, Xie Q, Chua NH (2003). The Arabidopsis auxin- inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951-1961. |
| [12] |
Huang TB, Irish VF (2016). Gene networks controlling petal organogenesis. J Exp Bot 67, 61-68.
DOI PMID |
| [13] | Kieber JJ, Schaller GE (2018). Cytokinin signaling in plant development. Development 145, dev149344. |
| [14] |
Krizek BA, Anderson JT (2013). Control of flower size. J Exp Bot 64, 1427-1437.
DOI PMID |
| [15] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402-408.
DOI PMID |
| [16] | Nag A, King S, Jack T (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106, 22534-22539. |
| [17] |
Nath U, Crawford BCW, Carpenter R, Coen E (2003). Genetic control of surface curvature. Science 299, 1404-1407.
DOI PMID |
| [18] | Nishijima T, Miyaki H, Sasaki K, Okazawa T (2006). Cultivar and anatomical analysis of corolla enlargement of petunia (Petunia hybrida Vilm.) by cytokinin application. Sci Hortic 111, 49-55. |
| [19] | Nishijima T, Niki T, Niki T (2011). The large-flowered petunia (Petunia hybrida Vilm.) genotype promotes expressions of type-A response regulator and cytokinin receptor genes like cytokinin response. J Japan Soc Hort Sci 80, 343-350. |
| [20] |
Santner A, Calderon-Villalobos LIA, Estelle M (2009). Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol 5, 301-307.
DOI PMID |
| [21] | Tan YQ, Yu HQ, Chen QS, Liu Y (2010). Application of excel in establishment of Chi-square test analysis module in agricultural statistic. Hubei Agricul Sci 49, 3192-3195. (in Chinese) |
| 谭永强, 余华强, 陈桥生, 刘莹 (2010). 利用Excel软件建立卡方检验分析模板在农业统计中的应用. 湖北农业科学 49, 3192-3195. | |
| [22] | To JPC, Deruère J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2007). Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19, 3901-3914. |
| [23] | To JPC, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658-671. |
| [24] |
Vandenbussche M, Chambrier P, Bento SR, Morel P (2016). Petunia, your next supermodel? Front Plant Sci 7, 72.
DOI PMID |
| [25] | Venail J, Dell'olivo A, Kuhlemeier C (2010). Speciation genes in the genus Petunia. Philos Trans R Soc Lond B Biol Sci 365, 461-468. |
| [26] | Verdonk JC, Shibuya K, Loucas HM, Colquhoun TA, Underwood BA, Clark DG (2008). Flower-specific expression of the Agrobacterium tumefaciens isopentenyl- transferase gene results in radial expansion of floral organs in Petunia hybrida. Plant Biotech J 6, 694-701. |
| [27] | Wang JT, Zhang YW, Du YW, Ren WL, Li HF, Sun WX, Ge C, Zhang YM (2022). SEA v2.0: an R software package for mixed major genes plus polygenes inheritance analysis of quantitative traits. Acta Agron Sin 48, 1416-1424. (in Chinese) |
|
王靖天, 张亚雯, 杜应雯, 任文龙, 李宏福, 孙文献, 葛超, 章元明 (2022). 数量性状主基因+多基因混合遗传分析R软件包SEA v2.0. 作物学报 48, 1416-1424.
DOI |
|
| [28] | Zhang ST, Zhou Q, Chen F, Wu L, Liu BJ, Li F, Zhang JQ, Bao MZ, Liu GF (2020). Genome-wide identification, characterization and expression analysis of TCP transcription factors in Petunia. Int J Mol Sci 21, 6594. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||