

植物学报 ›› 2018, Vol. 53 ›› Issue (4): 528-541.DOI: 10.11983/CBB18004 cstr: 32102.14.CBB18004
收稿日期:2018-01-03
接受日期:2018-03-29
出版日期:2018-07-01
发布日期:2018-09-11
通讯作者:
王影,邱丽娟
作者简介:† 共同第一作者。
Wang Ying1,2,*( ), Li Xianggan2, Qiu Lijuan1,*(
), Li Xianggan2, Qiu Lijuan1,*( )
)
Received:2018-01-03
Accepted:2018-03-29
Online:2018-07-01
Published:2018-09-11
Contact:
Wang Ying,Qiu Lijuan
About author:† These authors contributed equally to this paper
摘要: 近年来, CRISPR定点编辑技术发展迅猛, 在动物、植物和微生物中均得到广泛应用。其中, 备受关注的脱靶现象也是研究的热点, 迄今已取得了重要进展。该文介绍了脱靶现象的产生原理及体内和体外检测脱靶现象的方法, 评价了通过改进sgRNA设计和优化CRISPR系统等来降低脱靶率的方法。在植物基因组定点编辑过程中, 应适时检测脱靶现象, 提高脱靶检测的精确度和准确度。
王影, 李相敢, 邱丽娟. CRISPR/Cas9基因组定点编辑中脱靶现象的研究进展. 植物学报, 2018, 53(4): 528-541.
Wang Ying, Li Xianggan, Qiu Lijuan. Research Progress in Off-target in CRISPR/Cas9 Genome Editing. Chinese Bulletin of Botany, 2018, 53(4): 528-541.
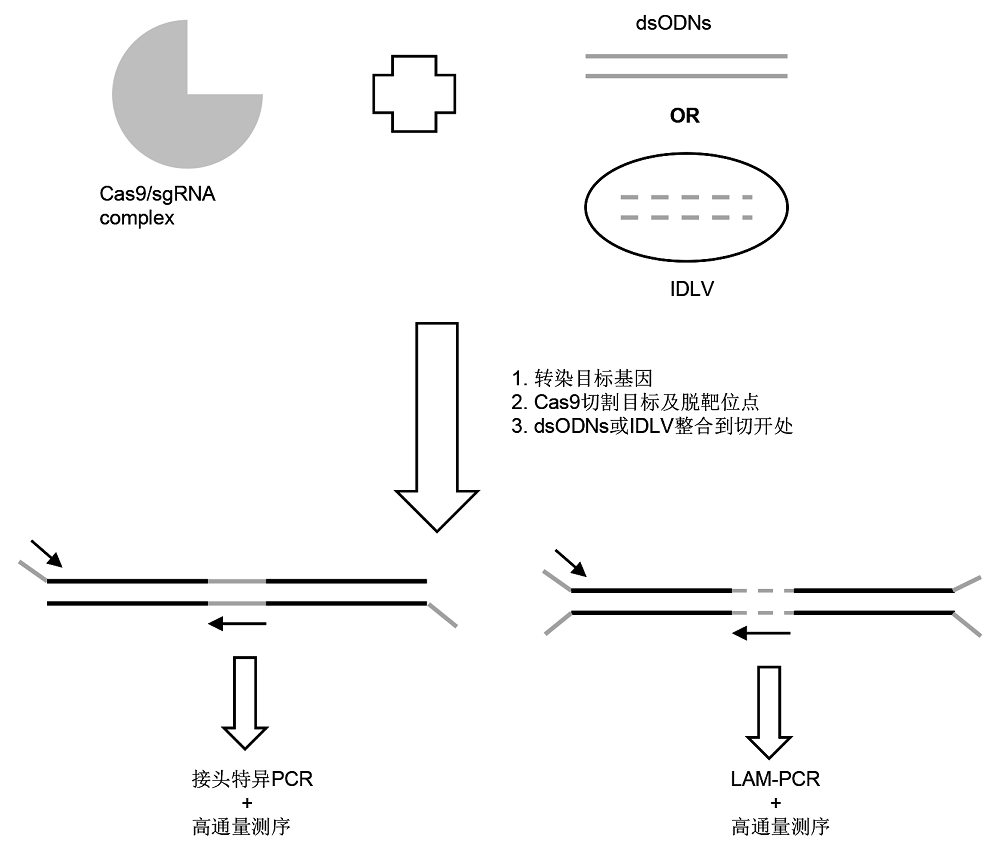
图1 GUIDE-seq和非整合病毒载体(IDLV)方法示意图
Figure 1 Schematic diagram of genome-wide unbiased identification of DSBs enabled by sequencing (GUIDE-seq) and integrase-defective lentiviral vectors (IDLV)
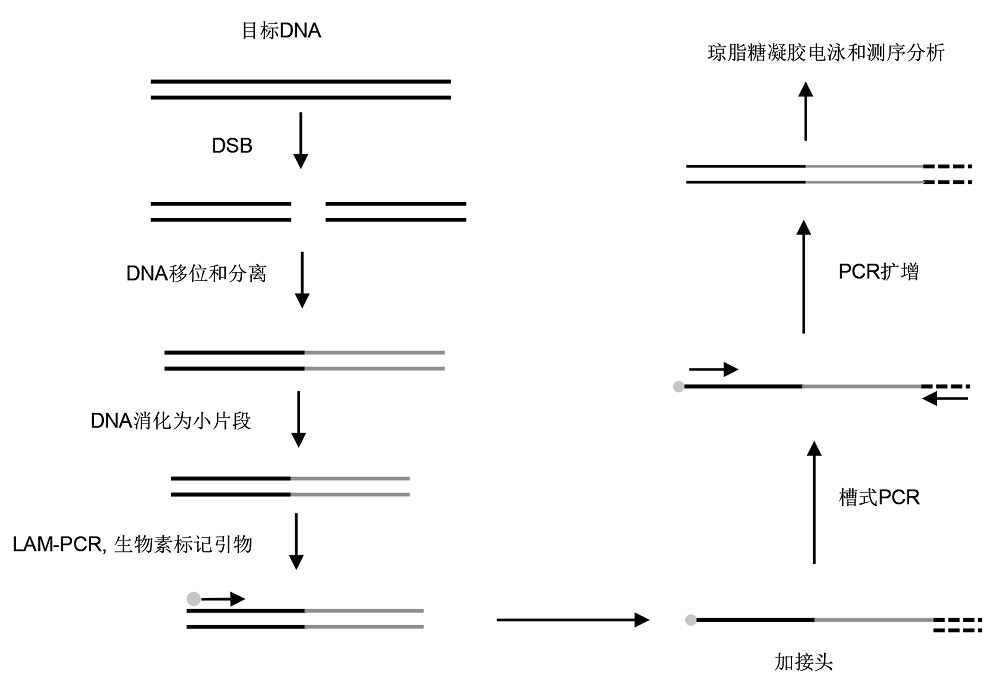
图2 高通量全基因组易位测序(HTGTS)的简化流程(改自Frock et al., 2015)
Figure 2 Simplified process of high-throughput genomewide translocation sequencing (HTGTS) (modified from Frock et al., 2015)
| 检测方法 | 优点 | 缺点 |
|---|---|---|
| 软件预测+测序 | 简单, 易操作 | 无法覆盖全基因组, 只检测软件预测的序列 |
| 深度测序 | 简单, 易操作 | 成本高, 数据分析复杂 |
| T7E1检测 | 成本低, 速度快 | 需辅助软件预测脱靶位点, 无法覆盖全基因组 |
| GUIDE-seq | 精确度高(0.1%), 全基因组检测 | 受细胞转染的限制, dsODNs的整合效率影响结果 |
| HTGTS | 全基因组覆盖 | 只能检测到与断裂片段结合的脱靶位点, 存在遗漏 |
| IDLV | 全基因组覆盖, 无偏见检测脱靶位点 | 精确度较低(1%) |
| BLESS | 摆脱了特定核酸酶的限制, 可检测任何酶所产生的DSB 中的突变 | 操作复杂, 只能检测特定时期所产生的突变 |
| ChIP-seq | 全基因组检测 | 体外检测, 未考虑切割频率, 准确度低 |
| Digenome-seq | 高效率, 高灵敏度(0.1%), 全基因组检测 | 体外检测, 成本高, 分析难度大 |
| Circle-seq | 全基因组检测, 灵敏度高, 提供分析平台 | 体外检测, 准确度不高 |
| FIND-seq | 全基因组检测, 灵敏度高 | 体外检测, 存在个别遗漏 |
| Site-seq | 全基因组检测, 灵敏度高 | 体外检测, 测序结果分析需要特定的算法 |
表1 脱靶的体内和体外检测方法对比
Table 1 Advantage and disadvantage of in vivo and in vitro off-target detection methods
| 检测方法 | 优点 | 缺点 |
|---|---|---|
| 软件预测+测序 | 简单, 易操作 | 无法覆盖全基因组, 只检测软件预测的序列 |
| 深度测序 | 简单, 易操作 | 成本高, 数据分析复杂 |
| T7E1检测 | 成本低, 速度快 | 需辅助软件预测脱靶位点, 无法覆盖全基因组 |
| GUIDE-seq | 精确度高(0.1%), 全基因组检测 | 受细胞转染的限制, dsODNs的整合效率影响结果 |
| HTGTS | 全基因组覆盖 | 只能检测到与断裂片段结合的脱靶位点, 存在遗漏 |
| IDLV | 全基因组覆盖, 无偏见检测脱靶位点 | 精确度较低(1%) |
| BLESS | 摆脱了特定核酸酶的限制, 可检测任何酶所产生的DSB 中的突变 | 操作复杂, 只能检测特定时期所产生的突变 |
| ChIP-seq | 全基因组检测 | 体外检测, 未考虑切割频率, 准确度低 |
| Digenome-seq | 高效率, 高灵敏度(0.1%), 全基因组检测 | 体外检测, 成本高, 分析难度大 |
| Circle-seq | 全基因组检测, 灵敏度高, 提供分析平台 | 体外检测, 准确度不高 |
| FIND-seq | 全基因组检测, 灵敏度高 | 体外检测, 存在个别遗漏 |
| Site-seq | 全基因组检测, 灵敏度高 | 体外检测, 测序结果分析需要特定的算法 |
| 设计软件 | 网址 | 软件功能 |
|---|---|---|
| Cas-OFFinder | http://www.rgenome.net/ | 针对CRISPR/Cas9系统, 通过使用者提供的目标位点序列, 推测所选择的目标基因组中潜在的脱靶位点。可选择不同的PAM、错配数量和是否允许错位配对( |
| CHOPCHOP | https://chopchop.rc.fas.harvard. edu/ | 针对CRISPR/Cas9和TALEN系统, 根据用户给出的目标基因序列, 在目标基因组或染色体中查找潜在的脱靶位点。可查找2个以内碱基错配的脱靶序列( |
| CRISPR Design | http://crispr.mit.edu/ | 在所给基因序列中设计sgRNA, 能够预测该sgRNA在基因组中的脱靶情况, 并标出最特异的sgRNA |
| CRISPR/Cas9 gRNA Finder | http://spot.colorado.edu/~slin/cas9.html | 在所给出的基因序列中, 查找可能的目标切割位点, 给出合适的sgRNA序列, 并推测其二级结构( |
| CRISPRfinder Christine | http://crispr.u-psud.fr/Server/ | 在公开的微生物基因组中定位CRISPR重复序列位置, 并能报告间隔序列( |
| E-CRISP | http://www.e-crisp.org/E-CRISP/ | 设计并评估CRISPR目标位点, 输入基因ID、FASTA序列进行搜索。可针对不同的CRISPR系统进行设计( |
| CRISPR-Plant | http://www.genome.arizona.edu/crispr/ | 此软件针对一系列植物基因组设计CRISPR目标位点, 在所给出的基因序列或染色体序列中查找合适的目标位点( |
| CRISPR MultiTargeter | http://www.multicrispr.net | 可用于设计同时靶向几个基因或1个基因中的多个位点的sgRNA ( |
| sgRNA Designer | http://www.broadinstitute.org/rnai/public/ | 适用于人类和小鼠基因组中sgRNA的设计。能够推荐最高特异性的sgRNA, 但不会给出可能的脱靶位点( |
| sgRNA Scorer | https://crispr.med.harvard.edu/sgRNAScorer/ | 可以设计sgRNA并评估sgRNA的体内切割活性( |
表2 sgRNA设计软件汇总
Table 2 Summary of sgRNA design tools
| 设计软件 | 网址 | 软件功能 |
|---|---|---|
| Cas-OFFinder | http://www.rgenome.net/ | 针对CRISPR/Cas9系统, 通过使用者提供的目标位点序列, 推测所选择的目标基因组中潜在的脱靶位点。可选择不同的PAM、错配数量和是否允许错位配对( |
| CHOPCHOP | https://chopchop.rc.fas.harvard. edu/ | 针对CRISPR/Cas9和TALEN系统, 根据用户给出的目标基因序列, 在目标基因组或染色体中查找潜在的脱靶位点。可查找2个以内碱基错配的脱靶序列( |
| CRISPR Design | http://crispr.mit.edu/ | 在所给基因序列中设计sgRNA, 能够预测该sgRNA在基因组中的脱靶情况, 并标出最特异的sgRNA |
| CRISPR/Cas9 gRNA Finder | http://spot.colorado.edu/~slin/cas9.html | 在所给出的基因序列中, 查找可能的目标切割位点, 给出合适的sgRNA序列, 并推测其二级结构( |
| CRISPRfinder Christine | http://crispr.u-psud.fr/Server/ | 在公开的微生物基因组中定位CRISPR重复序列位置, 并能报告间隔序列( |
| E-CRISP | http://www.e-crisp.org/E-CRISP/ | 设计并评估CRISPR目标位点, 输入基因ID、FASTA序列进行搜索。可针对不同的CRISPR系统进行设计( |
| CRISPR-Plant | http://www.genome.arizona.edu/crispr/ | 此软件针对一系列植物基因组设计CRISPR目标位点, 在所给出的基因序列或染色体序列中查找合适的目标位点( |
| CRISPR MultiTargeter | http://www.multicrispr.net | 可用于设计同时靶向几个基因或1个基因中的多个位点的sgRNA ( |
| sgRNA Designer | http://www.broadinstitute.org/rnai/public/ | 适用于人类和小鼠基因组中sgRNA的设计。能够推荐最高特异性的sgRNA, 但不会给出可能的脱靶位点( |
| sgRNA Scorer | https://crispr.med.harvard.edu/sgRNAScorer/ | 可以设计sgRNA并评估sgRNA的体内切割活性( |
| 1 | Bae S, Park J, Kim JS (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases.Bioinformatics 30, 1473-1475. |
| 2 | Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013). Plant genome editing made easy: targeted muta- genesis in model and crop plants using the CRISPR/Cas system.Plant Methods 9, 39. |
| 3 | Cameron P, Fuller CK, Donohoue PD, Jones BN, Thom- pson MS, Carter MM, Gradia S, Vidal B, Garner E, Slorach EM, Lau E, Banh LM, Lied AM, Edwards LS, Settle AH, Capurso D, Llaca V, Deschamps S, Cigan M, Young JK, May AP (2017). Mapping the genomic landscape of CRISPR-Cas9 cleavage.Nat Methods 14, 600-606. |
| 4 | Cencic R, Miura H, Malina A, Robert F, Ethier S, Schme- ing TM, Dostie J, Pelletier J (2014). Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage.PLoS One 9, e109213. |
| 5 | Chari R, Mali P, Moosburner M, Church GM (2015). Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach.Nat Methods 12, 823-826. |
| 6 | Chen XG, Lu XK, Shu N, Wang S, Wang JJ, Wang DL, Guo LX, Ye WW (2017). Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/Cas9 system. Sci Rep 7, 44304. |
| 7 | Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, Neuberg D, Monti S, Giallourakis CC, Gostissa M, Alt FW (2011). Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107-119. |
| 8 | Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS (2014). Analysis of off-target effects of CRISPR/Cas- derived RNA-guided endonucleases and nickases.Genome Res 24, 132-141. |
| 9 | Cradick TJ, Fine EJ, Antico CJ, Bao G (2013). CRISPR/ Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41, 9584-9592. |
| 10 | Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, Pasero P, Rowicka M, Dikic I (2013). Nucleotide-resolution DNA double-strand break mapping by next-generation sequen- cing.Nat Methods 10, 361-365. |
| 11 | Deveau H, Garneau JE, Moineau S (2010). CRISPR/Cas system and its role in phage-bacteria interactions.Annu Rev Microbiol 64, 475-493. |
| 12 | Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9.Nat Biotechnol 34, 184-191. |
| 13 | Duan JZ, Lu GQ, Xie Z, Lou ML, Luo J, Guo L, Zhang Y (2014). Genome-wide identification of CRISPR/Cas9 off- targets in human genome.Cell Res 24, 1009-1012. |
| 14 | Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, Kornepati AV, Sousa A, Collins MA, Jayaram H, Cullen BR, BumcrotEmail D (2015). Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol 16, 257. |
| 15 | Frock RL, Hu JZ, Meyers RM, Ho YJ, Kii E, Alt FW (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases.Nat Biotechnol 33, 179-186. |
| 16 | Fu YF, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013). High-frequency off-target mutag- enesis induced by CRISPR-Cas nucleases in human cells.Nat Biotechnol 31, 822-826. |
| 17 | Fu YF, Sander JD, Reyon D, Cascio VM, Joung JK (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs.Nat Biotechnol 32, 279-284. |
| 18 | Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang JB, Friedman G, Holmes MC, Gregory PD, Glimm H, Schmidt M, Naldini L, von Kalle C (2011). An unbiased genome-wide analysis of zinc-finger nuclease specificity.Nat Biotechnol 29, 816-823. |
| 19 | Harrington LB, Doxzen KW, Ma E, Liu JJ, Knott GJ, Edraki A, Garcia B, Amrani N, Chen JS, Cofsky JC, Kranzusch PJ, Sontheimer EJ, Davidson AR, Maxwell KL, Doudna JA (2017). A broad-spectrum inhibitor of CRISPR-Cas9.Cell 170, 1224-1233. |
| 20 | Havlicek S, Shen Y, Alpagu Y, Bruntraeger MB, Zufir NBM, Phuah ZY, Fu ZY, Dunn NR, Stanton LW (2017). Re-engineered RNA-guided fokI-nucleases for improved genome editing in human cells.Mol Ther 25, 342-355. |
| 21 | Hsu PD, Lander ES, Zhang F (2014). Development and applications of CRISPR-Cas9 for genome engineering.Cell 157, 1262-1278. |
| 22 | Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li YQ, Fine EJ, Wu XB, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F (2013). DNA targeting specificity of RNA-guided Cas9 nucleases.Nat Biotechnol 31, 827-832. |
| 23 | Jiang WY, Bikard D, Cox D, Zhang F, Marraffini LA (2013). RNA-guided editing of bacterial genomes using CRISPR- Cas systems.Nat Biotechnol 31, 233-239. |
| 24 | Jiang WZ, Yang B, Weeks DP (2014). Efficient CRISPR/ Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS One 9, e99225. |
| 25 | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.Science 337, 816-821. |
| 26 | Joung KJ, Tsai S (2017a). Full interrogation of nuclease DSBS and sequencing (find-seq). USA, 20170073747. 2017-03-16. |
| 27 | Joung KJ, Tsai S (2017b). Comprehensive in vitro reporting of cleavage events by sequencing (circle-seq). USA, 9850- 484. 2017-12-26. |
| 28 | Kaya H, Mikami M, Endo A, Endo M, Toki S (2016). Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci Rep 6, 26871. |
| 29 | Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS (2015). Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells.Nat Methods 12, 237-243. |
| 30 | Kim D, Kim S, Kim S, Park J, Kim JS (2016). Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq.Genome Res 26, 406-415. |
| 31 | Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng ZL, Joung JK (2016). High-fidelity CRISPR- Cas9 nucleases with no detectable genome-wide off-target effects.Nature 529, 490-495. |
| 32 | Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng ZL, Joung JK (2015a). Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 33, 1293-1298. |
| 33 | Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng ZL, Gonzales APW, Li ZY, Peterson RT, Yeh JRJ, Aryee MJ, Joung JK (2015b). Engineered CRISPR- Cas9 nucleases with altered PAM specificities.Nature 523, 481-485. |
| 34 | Kuscu C, Arslan S, Singh R, Thorpe J, Adli M (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease.Nat Biotechnol 32, 677-683. |
| 35 | Lee CM, Cradick TJ, Fine EJ, Bao G (2016). Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing.Mol Ther 24, 475-487. |
| 36 | Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, Lee K, Kim D, Kim S, Kim JS (2017). Directed evolution of CRISPR- Cas9 to increase its specificity.bioRxiv (1), 237040. |
| 37 | Li JC, Wang XB, Song WW, Huang XY, Zhou J, Zeng HY, Sun S, Jia HC, Li WB, Zhou XA, Li SZ, Chen PY, Wu CX, Guo Y, Han TF, Qiu LJ (2017). Genetic variation of maturity groups and four E genes in the Chinese soybean mini core collection. PLoS One 12, e0172106. |
| 38 | Li W, Teng F, Li TD, Zhou Q (2013). Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems.Nat Biotechnol 31, 684-686. |
| 39 | Lozano-Juste J, Cutler SR (2014). Plant genome engineering in full bloom.Trends Plant Sci 19, 284-287. |
| 40 | Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013). RNA-guided human genome engineering via Cas9.Science 339, 823-826. |
| 41 | Meng XB, Hu XX, Liu Q, Song XG, Gao CX, Li JY, Wang KJ (2018). Robust genome editing of CRISPR-Cas9 at NAG PAMs in rice.Sci China Life Sci 61, 122-125. |
| 42 | Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing.Nucleic Acids Res 42, W401-W407. |
| 43 | Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K (2016). Optimization of CRISPR/ Cas9 genome editing to modify abiotic stress responses in plants.Sci Rep 6, 26685. |
| 44 | Osborn MJ, Webber BR, Knipping F, Lonetree CL, Tennis N, DeFeo AP, McElroy AN, Starker CG, Lee C, Merkel S, Lund TC, Kelly-Spratt KS, Jensen MC, Voytas DF, von Kalle C, Schmidt M, Gabriel R, Hippen KL, Miller JS, Scharenberg AM, Tolar J, Blazar BR (2016). Eval- uation of TCR gene editing achieved by TALENs, CRISPR/ Cas9, and megaTAL nucleases.Mol Ther 24, 570-581. |
| 45 | Pan CT, Ye L, Qin L, Liu X, He YJ, Wang J, Chen LF, Lu G (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations.Sci Rep 6, 24765. |
| 46 | Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity.Nat Biotechnol 31, 839-843. |
| 47 | Peterson BA, Haak DC, Nishimura MT, Teixeira PJPL, James SR, Dangl JL, Nimchuk ZL (2016). Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis.PLoS One 11, e0162169. |
| 48 | Pourcel C, Drevet C (2013). Occurrence, diversity of CRISPR- Cas systems and genotyping implications. In: Barrangou R, van der Oost J, eds. CRISPR-Cas Systems. Berlin: Springer. pp. 33-59. |
| 49 | Prykhozhij SV, Rajan V, Gaston D, Berman JN (2015). CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences.PLoS One 10, e0119372. |
| 50 | Sampson TR, Weiss DS (2013). Alternative roles for CRISPR/Cas systems in bacterial pathogenesis.PLoS Pa- thog 9, e1003621. |
| 51 | Shen B, Zhang WS, Zhang J, Zhou JK, Wang JY, Chen L, Wang L, Hodgkins A, Iyer V, Huang XX, Skarnes WC (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects.Nat Methods 11, 399-402. |
| 52 | Shen CX, Que ZQ, Xia YM, Tang N, Li D, He RH, Cao ML (2017). Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J Plant Biol 60, 539-547. |
| 53 | Shin J, Jiang FG, Liu JJ, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA (2017). Disabling Cas9 by an anti-CRISPR DNA mimic.Sci Adv 3, e1701620. |
| 54 | Slaymaker IM, Gao LY, Zetsche B, Scott DA, Yan WX, Zhang F (2016). Rationally engineered Cas9 nucleases with improved specificity.Science 351, 84-88. |
| 55 | Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He CX, Wang Y, Brodsky RA, Zhang K, Cheng LZ, Ye ZH (2014). Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs.Cell Stem Cell 15, 12-13. |
| 56 | Tian SW, Jiang LJ, Gao Q, Zhang J, Zong M, Zhang HY, Ren Y, Guo SG, Gong GY, Liu F, Xu Y (2017). Efficient CRISPR/Cas9-based gene knockout in watermelon.Plant Cell Rep 36, 399-406. |
| 57 | Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK (2017). CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods 14, 607-614. |
| 58 | Tsai SQ, Zheng Zl, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases.Nat Biotechnol 33, 187-197. |
| 59 | Veres A, Gosis BS, Ding QR, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K (2014). Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing.Cell Stem Cell 15, 27-30. |
| 60 | Wang T, Wei JJ, Sabatini DM, Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system.Science 343, 80-84. |
| 61 | Wang XL, Wang YB, Wu XW, Wang JH, Wang YJ, Qiu ZJ, Chang T, Huang H, Lin RJ, Yee JK (2015). Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors.Nat Biotechnol 33, 175-178. |
| 62 | Wang Y, Geng LZ, Yuan ML, Wei J, Jin C, Li M, Yu K, Zhang Y, Jin HB, Wang E, Chai ZJ, Fu XD, Li XG (2017). Deletion of a target gene in indica rice via CRISPR/Cas9. Plant Cell Rep 36, 1333-1343. |
| 63 | Wu XB, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen SD, Jaenisch R, Zhang F, Sharp PA (2014). Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells.Nat Biotechnol 32, 670-676. |
| 64 | Xu C, Ren YH, Jian YQ, Guo ZF, Zhang Y, Xie CX, Fu JJ, Wang HW, Wang GY, Xu YB, Li P, Zou C (2017). Deve- lopment of a maize 55 K SNP array with improved genome coverage for molecular breeding.Mol Breed 37, 20. |
| 65 | Xu RF, Li H, Qin RY, Wang L, Li L, Wei PC, Yang JB (2014). Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 7, 5. |
| 66 | Zhang DB, Zhang HW, Li TD, Chen KL, Qiu JL, Gao CX (2017a). Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases.Genome Biol 18, 191. |
| 67 | Zhang H, Zhang JS, Wei PL, Zhang BT, Gou F, Feng ZY, Mao YF, Yang L, Zhang H, Xu NF, Zhu JK (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation.Plant Biotechnol J 12, 797-807. |
| 68 | Zhang Q, Xing HL, Wang ZP, Zhang HY, Yang F, Zhou Y, Wang XC, Chen QJ (2017b). High-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention by improving specificity of the tools.bioRxiv (1), 203489. |
| 69 | Zong Y, Wang YP, Li C, Zhang R, Chen KL, Ran YD, Qiu JL, Wang DW, Gao CX (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion.Nat Biotechnol 35, 438-440. |
| [1] | 苗春妍, 李铭铭, 左鑫, 丁宁, 杜家方, 李娟, 张重义, 王丰青. 湖北地黄CRISPR/Cas9基因编辑体系的建立[J]. 植物学报, 2023, 58(6): 905-916. |
| [2] | 何晓玲, 刘鹏程, 马伯军, 陈析丰. 基于CRISPR/Cas9的基因编辑技术研究进展及其在植物中的应用[J]. 植物学报, 2022, 57(4): 508-531. |
| [3] | 谢先荣, 曾栋昌, 谭健韬, 祝钦泷, 刘耀光. 基于CRISPR编辑系统的DNA片段删除技术[J]. 植物学报, 2021, 56(1): 44-49. |
| [4] | 谢卡斌. 中国科学家发现胞嘧啶单碱基编辑工具存在 基因组范围的脱靶[J]. 植物学报, 2019, 54(3): 296-299. |
| [5] | 苏钺凯,邱镜仁,张晗,宋振巧,王建华. CRISPR/Cas9系统在植物基因组编辑中技术改进与创新的研究进展[J]. 植物学报, 2019, 54(3): 385-395. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||