

植物学报 ›› 2024, Vol. 59 ›› Issue (3): 373-382.DOI: 10.11983/CBB23140 cstr: 32102.14.CBB23140
收稿日期:2023-10-15
接受日期:2024-01-02
出版日期:2024-05-10
发布日期:2024-05-10
通讯作者:
顾红雅, 北京大学生命科学学院教授, 博士生导师。长期从事植物遗传多样性和演化研究, 基因家族功能和演化研究。以通讯作者身份在Cell Research、Plant Cell、New Phytologist和PLoS Genetics等植物遗传及演化相关的国际期刊上发表研究论文80余篇; 参与编著教材3部, 科普专著2部, 翻译教材3部。目前其研究团队以拟南芥为模式生物, 收集了全国约10个省份几十份野生拟南芥居群, 利用遗传学、基因组学、群体遗传学和生物信息学等手段对其遗传多样性、演化历史及适应性机制进行研究。E-mail: 基金资助:
Jixuan Yang, Xuefei Wang, Hongya Gu*( )
)
Received:2023-10-15
Accepted:2024-01-02
Online:2024-05-10
Published:2024-05-10
Contact:
E-mail: 摘要: 开花时间是被子植物生活史中的关键节点。十字花科植物拟南芥(Arabidopsis thaliana)广布于世界各地, 在海拔4 000 m以上的青藏高原也发现了该物种的自然居群, 高原独特的环境塑造了其生活史的独特表型, 在开花时间上表现为中等程度早花。该研究构建了西藏拟南芥Lhasa居群的F2代作图群体, 基于全基因组测序的QTL-seq定位分析, 在该居群中定位到主效基因FLC, 并且鉴定到其第1个内含子中存在2 307 bp的缺失, 这种单倍型只存在于西藏拟南芥居群。利用CRISPR-Cas9技术构建了Lhasa背景的flc-/-突变体, 表现为开花时间显著提前。研究结果表明, 西藏拟南芥开花时间改变的主要原因是FLC第1个内含子缺失, 该变异并未使其丧失全部功能, 这可能有利于西藏拟南芥适应青藏高原独特的气候环境。
杨继轩, 王雪霏, 顾红雅. 西藏野生拟南芥开花时间变异的遗传基础. 植物学报, 2024, 59(3): 373-382.
Jixuan Yang, Xuefei Wang, Hongya Gu. Genetic Basis of Flowering Time Variations in Tibetan Arabidopsis thaliana. Chinese Bulletin of Botany, 2024, 59(3): 373-382.
| Ecotype and transgenic plant | Source |
|---|---|
| Col-0 | Arabidopsis Biological Resource Center |
| Lhasa | Collected from Lhasa, Tibet |
| Dagze | Collected from Dagze, Tibet |
| JXnfx | Collected from Nanfeng, Jiangxi |
| SXcgx | Collected from Chenggu, Shaanxi |
| CQtlx | Collected from Tongliang, Chongqing |
| flc-/- (Lhasa) | Constructed by this experiment |
表1 植物材料
Table 1 Plant materials used in this study
| Ecotype and transgenic plant | Source |
|---|---|
| Col-0 | Arabidopsis Biological Resource Center |
| Lhasa | Collected from Lhasa, Tibet |
| Dagze | Collected from Dagze, Tibet |
| JXnfx | Collected from Nanfeng, Jiangxi |
| SXcgx | Collected from Chenggu, Shaanxi |
| CQtlx | Collected from Tongliang, Chongqing |
| flc-/- (Lhasa) | Constructed by this experiment |
| Primer name | Primer sequence (5′-3′) |
|---|---|
| DT1-FLC-BsF | ATATATGGTCTCGATTGCCTTCTCCAAACGTCGCAAGTT |
| DT1-FLC-BsR | ATTATTGGTCTCGAAACCCGGCGATAAGTACGCCTTC |
| DT1-FLC-F0 | TGCCTTCTCCAAACGTCGCAAGTTTTAGAGCTAGAAATAGC |
| DT1-FLC-R0 | AACCCGGCGATAAGTACGCCTTCAATCTCTTAGTCGACTCTAC |
| FLC-RT-F | AACGTCGCAACGGTCTCA |
| FLC-RT-R | TCCCACAAGCTTGCTATCCA |
| TUB2-RT-F | GTTCTCGATGTTGTTCGTAAG |
| TUB2-RT-R | TGTAAGGCTCAACCACAGTAT |
表2 引物序列
Table 2 Primers used in this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| DT1-FLC-BsF | ATATATGGTCTCGATTGCCTTCTCCAAACGTCGCAAGTT |
| DT1-FLC-BsR | ATTATTGGTCTCGAAACCCGGCGATAAGTACGCCTTC |
| DT1-FLC-F0 | TGCCTTCTCCAAACGTCGCAAGTTTTAGAGCTAGAAATAGC |
| DT1-FLC-R0 | AACCCGGCGATAAGTACGCCTTCAATCTCTTAGTCGACTCTAC |
| FLC-RT-F | AACGTCGCAACGGTCTCA |
| FLC-RT-R | TCCCACAAGCTTGCTATCCA |
| TUB2-RT-F | GTTCTCGATGTTGTTCGTAAG |
| TUB2-RT-R | TGTAAGGCTCAACCACAGTAT |
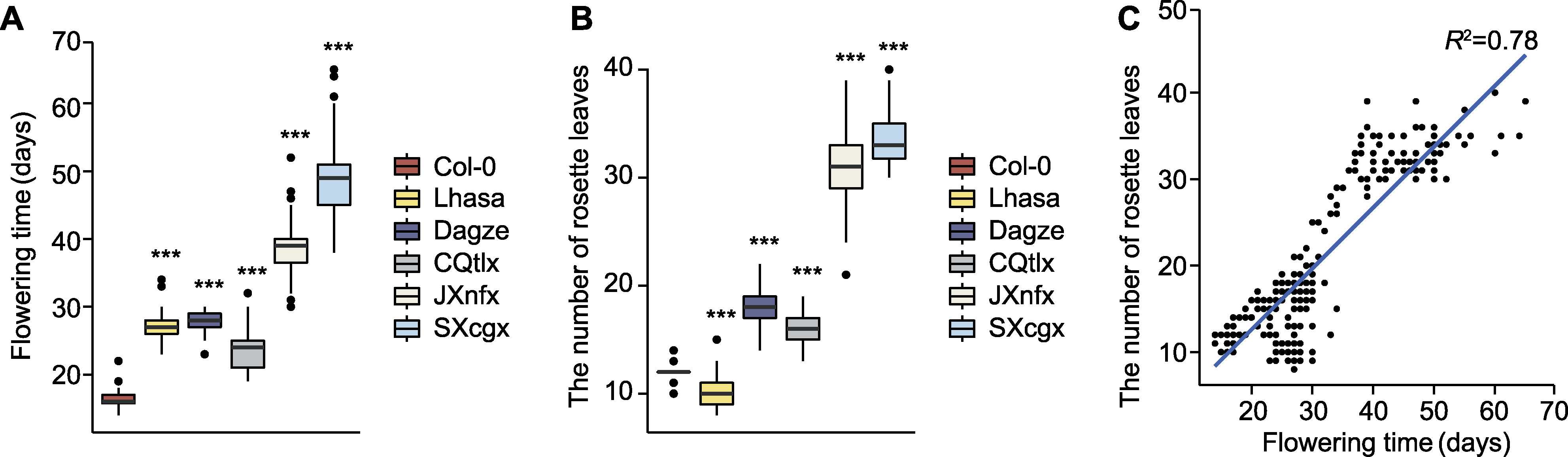
图1 西藏拟南芥和其它居群开花时间及莲座叶数目统计分析 (A) Col-0和部分中国野生居群开花时间统计(n≥40); (B) 开花时莲座叶数目统计(n≥40); (C) 开花时间与莲座叶数目的相关性分析。*** P<0.001 (Student’s t-test)。
Figure 1 Statistic analyses on the flowering time and number of rosette leaves of Tibetan and other Arabidopsis populations (A) Flowering time of Col-0 and some Chinese wild populations (n≥40); (B) Number of rosette leaves at flowering (n≥40); (C) Correlation analysis between flowering time and number of rosette leaves. *** P<0.001 (Student’s t-test).
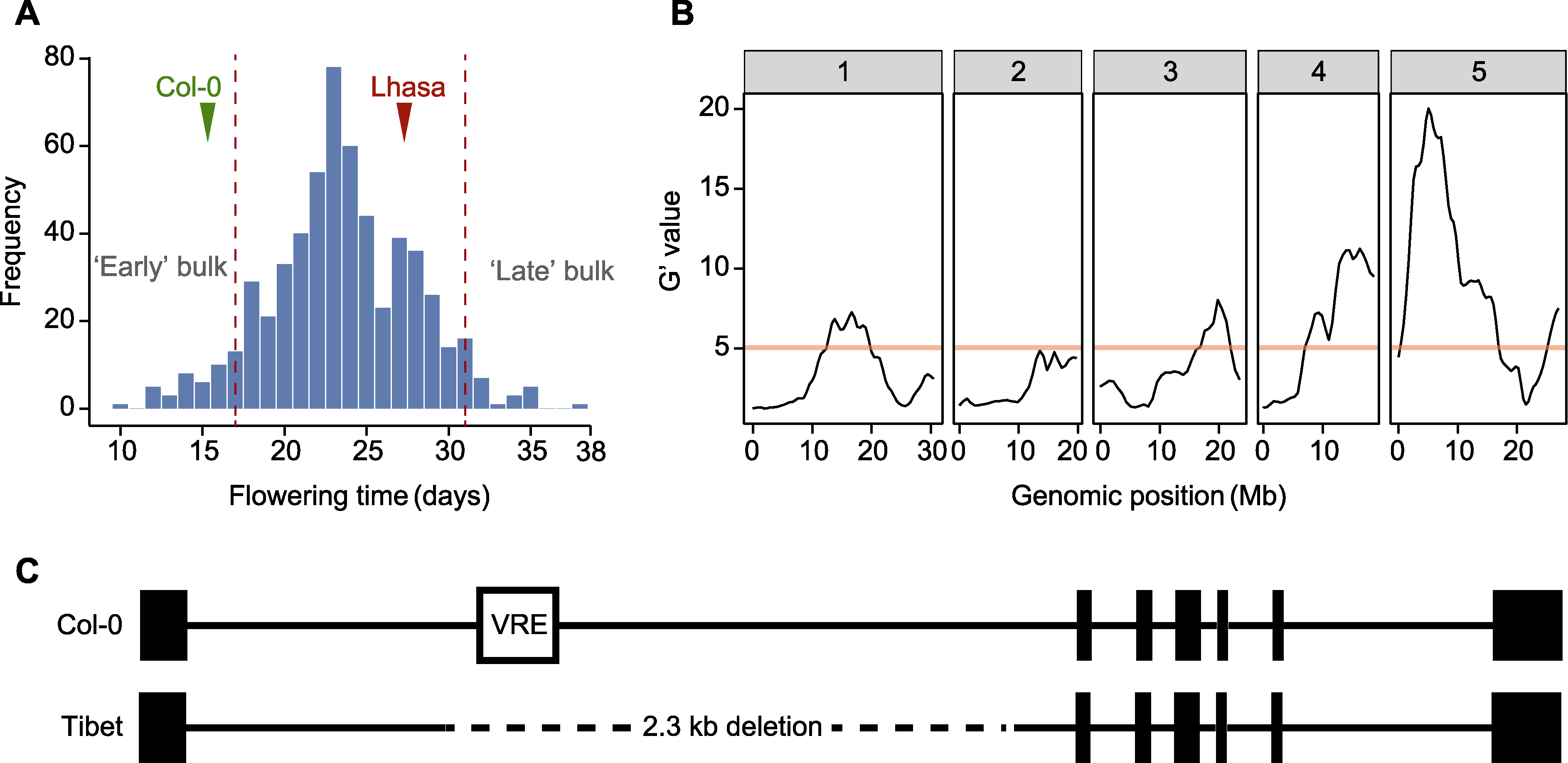
图2 F2代群体开花时间频率分布、QTL-seq定位结果和FLC基因序列变异 (A) F2代群体开花时间频率分布; (B) F2代群体QTL定位结果; (C) 西藏拟南芥FLC的变异, 黑色方块表示外显子, 黑色线段表示内含子。B图中的红色线段表示全基因组水平0.01的错误发现率。C图中的VRE表示春化响应元件, 虚线表示西藏居群中缺失的序列。
Figure 2 Frequency distribution of flowering time, QTL-seq mapping result in the F2 population and FLC gene sequence variation (A) Frequency distribution of flowering time in the F2 population; (B) QTL mapping result in the F2 population; (C) Variation in FLC of Tibetan Arabidopsis thaliana, black square indicates exon, and black line indicates intron. The genome-wide false discovery rate of 0.01 is indicated by the red line in Figure B. In Figure C, VRE indicates vernalization response element, and the dashed line indicates the deletion sequence in the Tibetan population.
| Population | Flowering time | Observed | Expected | χ2 | P-value |
|---|---|---|---|---|---|
| Lhasa-F2 | Early (<26 days) | 428 | 432 | 0.15 | 0.70 |
| Late (≥27 days) | 148 | 144 | |||
| Total | 576 | 576 |
表3 F2代群体开花时间分布的卡方检验
Table 3 Chi-square test for the distribution of flowering time in F2 populations
| Population | Flowering time | Observed | Expected | χ2 | P-value |
|---|---|---|---|---|---|
| Lhasa-F2 | Early (<26 days) | 428 | 432 | 0.15 | 0.70 |
| Late (≥27 days) | 148 | 144 | |||
| Total | 576 | 576 |
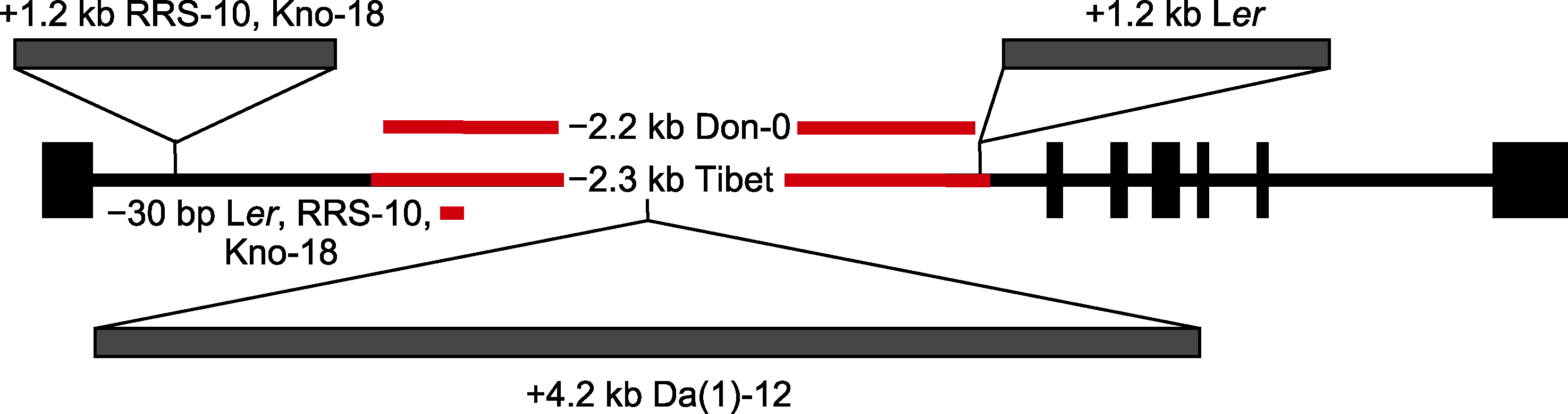
图3 部分拟南芥居群FLC第1个内含子区域的变异(改自Michaels et al., 2003; Strange et al., 2011; Méndez-Vigo et al., 2016) 以Col-0的FLC基因为参考, 黑色方形表示外显子, 黑色线段表示内含子, 红色线段表示序列缺失, 灰色方形表示序列插入。
Figure 3 Variation in the first intron of FLC in some Arabidopsis populations (modified from Michaels et al., 2003; Strange et al., 2011; Méndez-Vigo et al., 2016) FLC gene of Col-0 as a reference, black rectangle indicates exon, and black line indicates intron, red line indicates sequence deletion, and gray rectangle indicates sequence insertion.
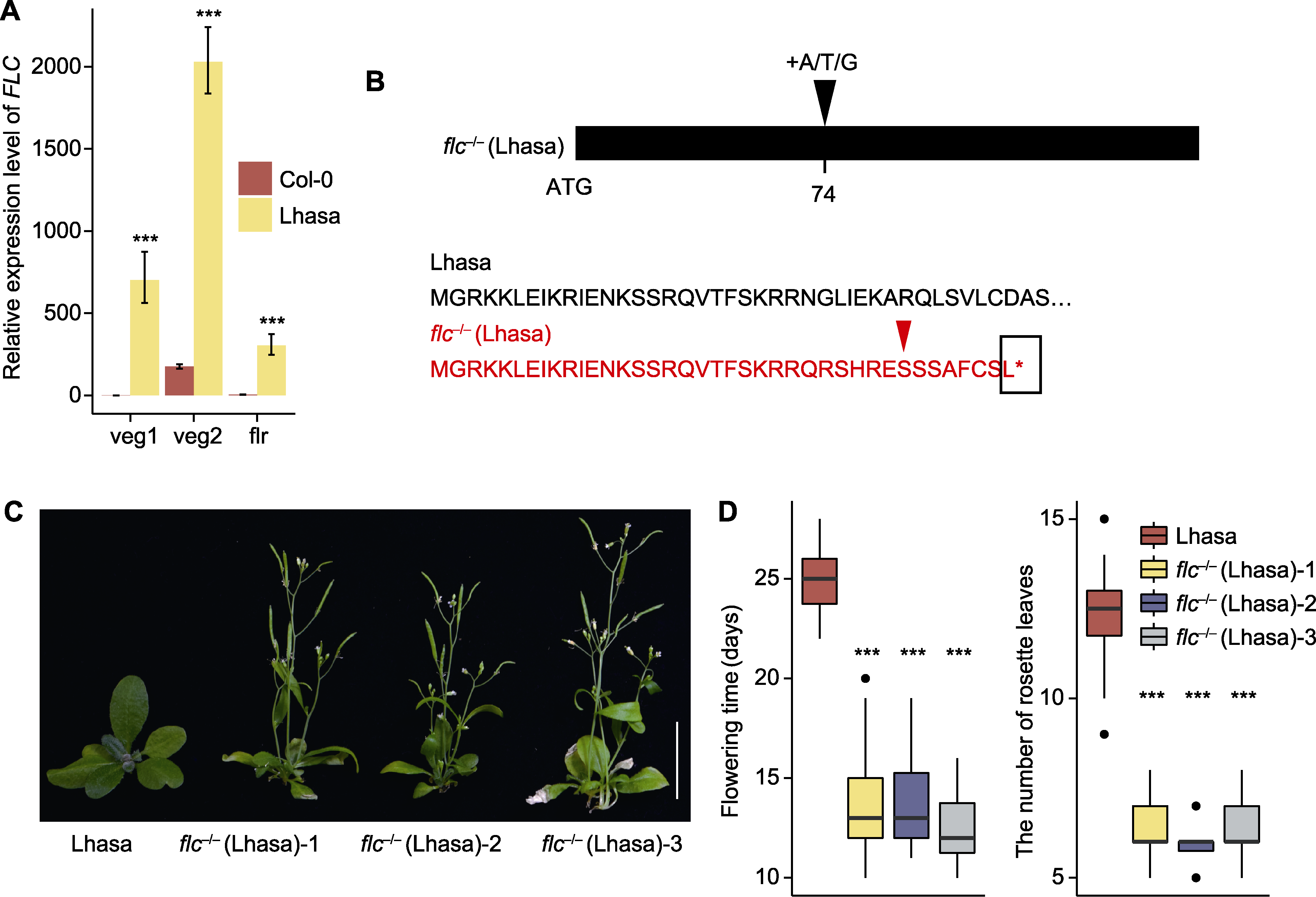
图4 Lhasa居群和flc-/- (Lhasa)突变体的开花表型、FLC的表达模式及flc-/- (Lhasa)的序列变异 (A) Col-0和Lhasa居群不同发育时期FLC的相对表达水平(veg1和veg2分别表示长出第5和第10片莲座叶时期, flr表示开花时期); (B) Lhasa居群和flc-/- (Lhasa)突变体的DNA和氨基酸序列变异形式(74表示自ATG开始的第74位碱基, 红色序列表示flc-/- (Lhasa)突变体的氨基酸序列, 红色箭头表示氨基酸开始变异的位点, 黑色方框表示氨基酸的终止位点); (C) Lhasa居群和flc-/- (Lhasa)突变体的开花表型(bar=2 cm), 拍摄于幼苗移栽后25天; (D) Lhasa居群和flc-/- (Lhasa)突变体的开花时间和开花时莲座叶数目(n≥40)。*** P<0.001 (Student's t-test)。
Figure 4 Flowering phenotypes of Lhasa population and flc-/- (Lhasa) mutants, FLC expression pattern, and sequence variation of flc-/- (Lhasa) (A) Relative expression levels of FLC at different development stages in Col-0 and Lhasa population (veg1 and veg2 indicate the time of growing the 5th and 10th rosette leaves, respectively, and flr indicating the time of flowering); (B) Variant forms of DNA and amino acid sequences of Lhasa population and flc-/- (Lhasa) mutants (74 indicates the 74th base from ATG, red sequence indicates the amino acid sequence of flc-/- (Lhasa) mutants, red arrow indicates site where amino acid begin to change in the flc-/- (Lhasa) mutants and black box indicates the termination site in flc-/- (Lhasa) mutants); (C) Flowering phenotypes of Lhasa population and flc-/- (Lhasa) mutants (bar=2 cm), photographed 25 d after seedling transplantation; (D) Flowering time and number of rosette leaves at flowering in Lhasa population and flc-/- (Lhasa) mutants (n≥40). *** P<0.001 (Student’s t-test).
| [1] | Adamczyk BJ, Lehti-Shiu MD, Fernandez DE (2007). The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50, 1007-1019. |
| [2] | Angel A, Song J, Dean C, Howard M (2011). A polycomb- based switch underlying quantitative epigenetic memory. Nature 476, 105-108. |
| [3] | Angel A, Song J, Yang HC, Questa JI, Dean C, Howard M (2015). Vernalizing cold is registered digitally at FLC. Proc Natl Acad Sci USA 112, 4146-4151. |
| [4] | Berry S, Hartley M, Olsson TSG, Dean C, Howard M (2015). Local chromatin environment of a polycomb target gene instructs its own epigenetic inheritance. eLife 4, e07205. |
| [5] | Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289-303. |
| [6] |
Crevillén P, Dean C (2011). Regulation of the floral repressor gene FLC: the complexity of transcription in a chromatin context. Curr Opin Plant Biol 14, 38-44.
DOI PMID |
| [7] | DeLeo VL, Menge DNL, Hanks EM, Juenger TE, Lasky JR (2020). Effects of two centuries of global environmental variation on phenology and physiology of Arabidopsis thaliana. Glob Change Biol 26, 523-538. |
| [8] | Ellis TJ, Postma FM, Oakley CG, Ågren J (2021). Life-history trade-offs and the genetic basis of fitness in Arabidopsis thaliana. Mol Ecol 30, 2846-2858. |
| [9] |
He F, Kang D, Ren Y, Qu LJ, Zhen Y, Gu H (2007). Genetic diversity of the natural populations of Arabidopsis thaliana in China. Heredity 99, 423-431.
PMID |
| [10] |
Heo JB, Sung S (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76-79.
DOI PMID |
| [11] | Hepworth J, Antoniou-Kourounioti RL, Berggren K, Selga C, Tudor EH, Yates B, Cox D, Collier Harris BR, Irwin JA, Howard M, Säll T, Holm S, Dean C (2020). Natural variation in autumn expression is the major adaptive determinant distinguishing Arabidopsis FLC haplotypes. eLife 9, e57671. |
| [12] | Jiang DH, Gu XF, He YH (2009). Establishment of the winte- rannual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21, 1733-1746. |
| [13] |
Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344-347.
DOI PMID |
| [14] | Jung JH, Lee HJ, Ryu JY, Park CM (2016). SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol Plant 9, 1647-1659. |
| [15] | Kim DH, Sung S (2017). Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev Cell 40, 302-312. |
| [16] |
Kinoshita A, Richter R (2020). Genetic and molecular basis of floral induction in Arabidopsis thaliana. J Exp Bot 71, 2490-2504.
DOI PMID |
| [17] | Koornneef M, Hanhart CJ, van der Veen JH (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229, 57-66. |
| [18] | Krämer U (2015). Planting molecular functions in an ecological context with Arabidopsis thaliana. eLife 4, e06100. |
| [19] | Le Corre V, Roux F, Reboud X (2002). DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19, 1261-1271. |
| [20] | Lee I, Bleecker A, Amasino R (1993). Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol Gen Genet 237, 171-176. |
| [21] |
Li H, Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-1760.
DOI PMID |
| [22] | Li PJ, Filiault D, Box MS, Kerdaffrec E, van Oosterhout C, Wilczek AM, Schmitt J, McMullan M, Bergelson J, Nordborg M, Dean C (2014). Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev 28, 1635-1640. |
| [23] | Liu ZW, Zhao N, Su YN, Chen SS, He XJ (2020). Exogenously overexpressed intronic long noncoding RNAs activate host gene expression by affecting histone modification in Arabidopsis. Sci Rep 10, 3094. |
| [24] | Mansfeld BN, Grumet R (2018). QTLseqr: an R package for bulk segregant analysis with next-generation sequencing. Plant Genome 11, 180006. |
| [25] |
Marquardt S, Boss PK, Hadfield J, Dean C (2006). Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. J Exp Bot 57, 3379-3386.
DOI PMID |
| [26] |
Martínez-Berdeja A, Stitzer MC, Taylor MA, Okada M, Ezcurra E, Runcie DE, Schmitt J (2020). Functional variants of DOG1 control seed chilling responses and variation in seasonal life-history strategies in Arabidopsis thaliana. Proc Natl Acad Sci USA 117, 2526-2534.
DOI PMID |
| [27] |
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297-1303.
DOI PMID |
| [28] | Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C (2011). Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol 157, 1942-1955. |
| [29] | Méndez-Vigo B, Savic M, Ausín I, Ramiro M, Martín B, Picó FX, Alonso-Blanco C (2016). Environmental and genetic interactions reveal FLOWERING LOCUS C as a modulator of the natural variation for the plasticity of flowering in Arabidopsis. Plant Cell Environ 39, 282-294. |
| [30] |
Michaels SD, He YH, Scortecci KC, Amasino RM (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100, 10102-10107.
PMID |
| [31] | Mitchell-Olds T, Schmitt J (2006). Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441, 947-952. |
| [32] |
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847-857.
DOI PMID |
| [33] | Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138, 1163-1173. |
| [34] | Shindo C, Bernasconi G, Hardtke CS (2007). Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Ann Bot 99, 1043-1054. |
| [35] | Springthorpe V, Penfield S (2015). Flowering time and seed dormancy control use external coincidence to generate life history strategy. eLife 4, e05557. |
| [36] | Strange A, Li PJ, Lister C, Anderson J, Warthmann N, Shindo C, Irwin J, Nordborg M, Dean C (2011). Majo- reffect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS One 6, e19949. |
| [37] | Sung S, He YH, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM (2006). Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38, 706-710. |
| [38] | Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013). QTL- seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74, 174-183. |
| [39] | The 1001 Genomes Consortium (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481-491. |
| [40] | Xie YR, Zhou Q, Zhao YP, Li QQ, Liu Y, Ma MD, Wang BB, Shen RX, Zheng ZG, Wang HY (2020). FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate Arabidopsis flowering. Mol Plant 13, 483-498. |
| [41] | Xu XR, Xu JY, Yuan C, Chen QQ, Liu QG, Wang XM, Qin C (2022). BBX17 interacts with CO and negatively regulates flowering time in Arabidopsis thaliana. Plant Cell Physiol 63, 401-409. |
| [42] | Yin P, Kang JQ, He F, Qu LJ, Gu HY (2010). The origin of populations of Arabidopsis thaliana in China, based on the chloroplast DNA sequences. BMC Plant Biol 10, 22. |
| [43] | Zan YJ, Carlborg Ö (2019). A polygenic genetic architecture of flowering time in the worldwide Arabidopsis thaliana population. Mol Biol Evol 36, 141-154. |
| [44] | Zeng LY, Gu ZY, Xu M, Zhao N, Zhu WD, Yonezawa T, Liu TM, Qiong L, Tersing T, Xu LL, Zhang Y, Xu RY, Sun NY, Huang YY, Lei JK, Zhang L, Xie F, Zhang F, Gu HY, Geng YP, Hasegawa M, Yang ZH, Crabbe MJC, Chen F, Zhong Y (2017). Discovery of a high-altitude ecotype and ancient lineage of Arabidopsis thaliana from Tibet. Sci Bull 62, 1628-1630. |
| [45] | Zhang YN, Zhou YP, Chen QH, Huang XL, Tian CE (2014). Molecular basis of flowering time regulation in Arabidopsis. Chin Bull Bot 49, 469-482. (in Chinese) |
|
张艺能, 周玉萍, 陈琼华, 黄小玲, 田长恩 (2014). 拟南芥开花时间调控的分子基础. 植物学报 49, 469-482.
DOI |
|
| [46] | Zhu P, Lister C, Dean C (2021). Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature 599, 657-661. |
| [47] | Zou YP, Hou XH, Wu Q, Chen JF, Li ZW, Han TS, Niu XM, Yang L, Xu YC, Zhang J, Zhang FM, Tan DY, Tian ZX, Gu HY, Guo YL (2017). Adaptation of Arabidopsis thaliana to the Yangtze River basin. Genome Biol 18, 239. |
| [1] | 景昭阳, 程可光, 舒恒, 马永鹏, 刘平丽. 全基因组重测序方法在濒危植物保护中的应用[J]. 生物多样性, 2023, 31(5): 22679-. |
| [2] | 杨小凤, 李小蒙, 廖万金. 植物开花时间的遗传调控通路研究进展[J]. 生物多样性, 2021, 29(6): 825-842. |
| [3] | 王蒙, 王婷, 夏增强, 李廷章, 金效华, 严岳鸿, 陈建兵. 基于转录组数据揭示4种兜兰的全基因组复制历史[J]. 植物学报, 2021, 56(6): 699-714. |
| [4] | 张长生, 魏滔, 周玉萍, 范甜, 吕天晓, 田长恩. FLC调控植物成花的分子机制研究新进展[J]. 植物学报, 2021, 56(6): 651-663. |
| [5] | 王婷, 夏增强, 舒江平, 张娇, 王美娜, 陈建兵, 王慷林, 向建英, 严岳鸿. 全基因组复制事件的绝对定年揭示莲座蕨属植物的迟滞演化[J]. 生物多样性, 2021, 29(6): 722-734. |
| [6] | 李琳, 路宁娜, 樊宝丽, 赵志刚. 雌雄异熟植物露蕊乌头开花时间对雌雄功能期及表型性别的影响[J]. 生物多样性, 2016, 24(6): 665-671. |
| [7] | 张艺能, 周玉萍, 陈琼华, 黄小玲, 田长恩. 拟南芥开花时间调控的分子基础[J]. 植物学报, 2014, 49(4): 469-482. |
| [8] | 刘乐乐, 刘左军, 杜国祯, 赵志刚. 毛茛状金莲花不同花期的花特征和访花昆虫的变化及表型选择[J]. 生物多样性, 2012, 20(3): 317-323. |
| [9] | 罗睿*;郭建军. 植物开花时间: 自然变异与遗传分化[J]. 植物学报, 2010, 45(01): 109-118. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||