

植物学报 ›› 2022, Vol. 57 ›› Issue (1): 1-11.DOI: 10.11983/CBB21183 cstr: 32102.14.CBB21183
• 特邀综述 • 下一篇
收稿日期:2021-10-25
接受日期:2022-01-09
出版日期:2022-01-01
发布日期:2022-01-17
通讯作者:
熊延
作者简介:* E-mail: yanxiong@fafu.edu.cn†共同第一作者。
基金资助:
Yanyan Meng1,2†, Nan Zhang1,2†, Yan Xiong1,2,*( )
)
Received:2021-10-25
Accepted:2022-01-09
Online:2022-01-01
Published:2022-01-17
Contact:
Yan Xiong
About author:First author contact:†These authors contributed equally to this paper.
摘要: 雷帕霉素靶蛋白(TOR)是真核生物中高度保守的丝氨酸/苏氨酸蛋白激酶, 能整合营养、能量、生长因子及环境信号, 协调细胞增殖、生长和代谢等过程, 是真核生物生长发育的核心调控因子。近年来, 随着相关研究系统的建立, 植物TOR的功能和机制研究取得了众多突破, 发现其进化上保守的生物学功能及植物中特有的信号通路。该文概述了TOR蛋白复合体的构成, 以及植物TOR响应糖、营养元素(氮、磷和硫)、激素及逆境胁迫信号来调控下游基因转录、蛋白翻译、代谢、细胞自噬和胁迫应答等生物学过程的分子机制, 并提出了植物TOR领域一些亟待解决的科学问题, 以期为全面揭示植物TOR的生物学功能提供参考。
孟彦彦, 张楠, 熊延. 植物TOR激酶响应上游信号的研究进展. 植物学报, 2022, 57(1): 1-11.
Yanyan Meng, Nan Zhang, Yan Xiong. Novel Links in the Plant Target of Rapamycin Signaling Networks. Chinese Bulletin of Botany, 2022, 57(1): 1-11.
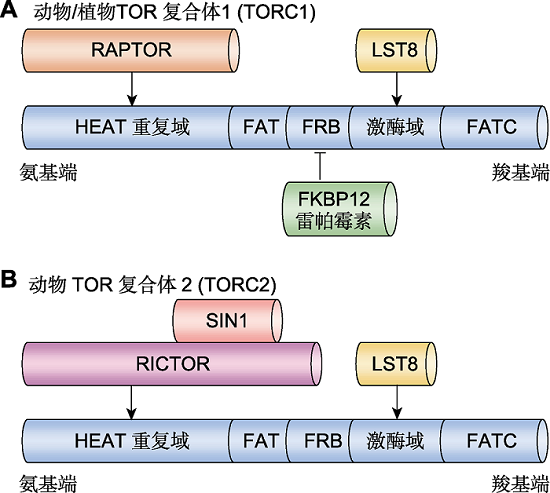
图1 动、植物中TOR蛋白的结构域和复合体组分 (A) 动、植物中TOR复合体1 (TORC1)。TORC1主要包含TOR、RAPTOR及LST8三个组分。动、植物中的TOR蛋白高度保守, 均由HEAT重复域、FAT、FRB、激酶域及FATC五个结构域组成。其中, HEAT重复域与RAPTOR结合; 激酶域与LST8结合; FRB结构域可被FKBP12介导与雷帕霉素结合。(B) 动物中TOR复合体2 (TORC2)。动物TORC2主要包含TOR、RICTOR、SIN1及LST8四个组分。其中RICTOR与SIN1互作并与HEAT重复域结合, LST8同样与激酶域结合。而尚未在植物中鉴定到RICTOR和SIN1的同源基因, 暗示TORC2在植物中可能不保守。
Figure 1 The domain structures and components of TOR complexes in plants and animals (A) TOR complex 1 (TORC1) in plants and animals. TORC1 mainly contains TOR, RAPTOR and LST8. TOR protein is highly conserved among plants and animals, composed of HEAT repeats, FAT, FRB, kinase and FATC domains. RAPTOR binds to the HEAT repeats, LST8 binds to the kinase domain, and FKBP12 mediates interact between the FRB domain and rapamycin. (B) TOR complex 2 (TORC2) in animals. TORC2 mainly contains TOR, RICTOR, SIN1 and LST8 in animal. RICTOR interacts with SIN1 and binds to HEAT repeats, and LST8 binds to the kinase domain. Either RICTOR or SIN1 has no homologous genes in plants, which indicates that TORC2 is not conserved.
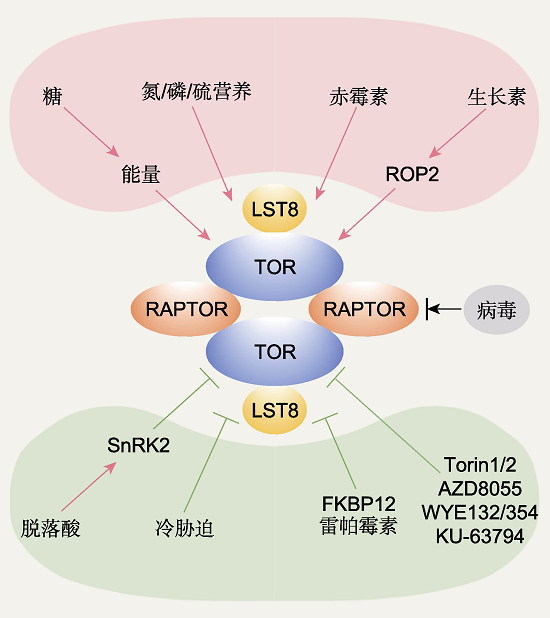
图2 植物TOR复合体1的上游调控因子 植物TOR复合体1感知多重上游信号, 并可能以二聚体的形式发挥作用。其中糖-能量信号、氮/磷/硫营养信号、生长素-ROP2信号以及赤霉素信号是TOR上游的激活信号; 脱落酸-SnRK2信号和冷胁迫信号是TOR上游的抑制信号; 不同病毒对TOR的作用不同, 既有促进也有抑制。FKBP12-雷帕霉素和ATP竞争性抑制剂Torin1/2、AZD8055、WYE132/354以及KU-63794均可特异且高效地抑制TOR激酶活性。红色箭头代表激活作用, 绿色T形线条代表抑制作用, 黑色线条代表兼具激活和抑制作用。
Figure 2 The upstream regulators of TOR complex 1 (TORC1) in plants TORC1 in plants senses multiple upstream signals, and probably functions as a dimerization form. Glucose-energy, nitrogen/phosphate/sulfur, auxin-ROP2, and GA signals are positive upstream regulators of TOR; ABA-SnRK2 and cold stress are negative upstream regulators of TOR; viruses play both positive and negative roles on TOR activity. FKBP12- rapamycin and ATP competitive inhibitors Torin1/2, AZD8055, WYE132/354 and KU-63794 inhibit plant TOR kinase activity specifically and efficiently. The red arrows represent activation, the green T shape lines mean repression, and the black line represents both activation and repression exist.
| [1] |
陈唯, 曾晓贤, 谢楚萍, 田长恩, 周玉萍 (2019). 植物内源ABA水平的动态调控机制. 植物学报 54, 677-687.
DOI |
| [2] | 温兴, 晋莲, 郭红卫 (2021). 甜蜜的相遇--营养与激素信号协同调节植物生长的新机制. 植物学报 56, 138-141. |
| [3] |
Albert V, Hall MN (2015). mTOR signaling in cellular and organismal energetics. Curr Opin Cell Biol 33, 55-66.
DOI URL |
| [4] |
Anderson GH, Veit B, Hanson MR (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol 3, 12.
PMID |
| [5] |
Aylett CHS, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T (2016). Architecture of human mTOR com- plex 1. Science 351, 48-52.
DOI URL |
| [6] |
Beltrán-Peña E, Aguilar R, Ortíz-López A, Dinkova TD, De Jiménez ES (2002). Auxin stimulates S6 ribosomal protein phosphorylation in maize thereby affecting protein synthesis regulation. Physiol Plant 115, 291-297.
PMID |
| [7] |
Brunkard JO (2020). Exaptive evolution of target of rapamycin signaling in multicellular eukaryotes. Dev Cell 54, 142-155.
DOI PMID |
| [8] |
Brunkard JO, Xu M, Scarpin MR, Chatterjee S, Shemyakina EA, Goodman HM, Zambryski P (2020). TOR dynamically regulates plant cell-cell transport. Proc Natl Acad Sci USA 117, 5049-5058.
DOI URL |
| [9] |
Burkart GM, Brandizzi F (2021). A tour of TOR complex signaling in plants. Trends Biochem Sci 46, 417-428.
DOI PMID |
| [10] |
Busche M, Scarpin MR, Hnasko R, Brunkard JO (2021). TOR coordinates nucleotide availability with ribosome biogenesis in plants. Plant Cell 33, 1615-1632.
DOI URL |
| [11] |
Cao PF, Kim SJ, Xing AQ, Schenck CA, Liu L, Jiang N, Wang J, Last RL, Brandizzi F (2019). Homeostasis of branched-chain amino acids is critical for the activity of TOR signaling in Arabidopsis. eLife 8, e50747.
DOI URL |
| [12] |
Chen GH, Liu MJ, Xiong Y, Sheen J, Wu SH (2018). TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc Natl Acad Sci USA 115, 12823-12828.
DOI URL |
| [13] |
Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y (2020). Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62, 25-54.
DOI URL |
| [14] |
Couso I, Pérez-Pérez ME, Ford MM, Martínez-Force E, Hicks LM, Umen JG, Crespo JL (2020). Phosphorus availability regulates TORC1 signaling via LST8 in chlamydomonas. Plant Cell 32, 69-80.
DOI URL |
| [15] |
David-Morrison G, Xu Z, Rui YN, Charng WL, Jaiswal M, Yamamoto S, Xiong B, Zhang K, Sandoval H, Duraine L, Zuo ZY, Zhang S, Bellen HJ (2016). WAC regulates mTOR activity by acting as an adaptor for the TTT and Pontin/Reptin complexes. Dev Cell 36, 139-151.
DOI PMID |
| [16] |
De Vleesschauwer D, Filipe O, Hoffman G, Seifi HS, Haeck A, Canlas P, Van Bockhaven J, De Waele E, Demeestere K, Ronald P, Hofte M (2018). Target of rapamycin signaling orchestrates growth-defense trade-offs in plants. New Phytol 217, 305-319.
DOI PMID |
| [17] |
Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8, 864-870.
DOI URL |
| [18] |
Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C (2016). TOR signaling and nutrient sensing. Annu Rev Plant Biol 67, 261-285.
DOI PMID |
| [19] |
Dong Y, Teleman AA, Jedmowski C, Wirtz M, Hell R (2019). The Arabidopsis THADA homologue modulates TOR activity and cold acclimation. Plant Biol 21, 77-83.
DOI URL |
| [20] |
Dong YH, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Samami AA, Wanatabe M, Sticht C, Teleman AA, Deragon JM, Saito K, Hell R, Wirtz M (2017). Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8, 1174.
DOI URL |
| [21] |
Fu LW, Liu YL, Qin GC, Wu P, Zi HL, Xu ZT, Zhao XD, Wang Y, Li YX, Yang SH, Peng C, Wong CCL, Yoo SD, Zuo ZC, Liu RY, Cho YH, Xiong Y (2021). The TOR- EIN2 axis mediates nuclear signaling to modulate plant growth. Nature 591, 288-292.
DOI URL |
| [22] |
Fu LW, Wang PC, Xiong Y (2020). Target of rapamycin signaling in plant stress responses. Plant Physiol 182, 1613-1623.
DOI URL |
| [23] |
Garber K (2001). Rapamycin may prevent post-transplant lymphoma. J Natl Cancer Inst 93, 1519.
PMID |
| [24] |
Garcia N, Li YB, Dooner HK, Messing J (2017). Maize defective kernel mutant generated by insertion of a Ds element in a gene encoding a highly conserved TTI2 cochaperone. Proc Natl Acad Sci USA 114, 5165-5170.
DOI URL |
| [25] |
González A, Hall MN, Lin SC, Hardie DG (2020). AMPK and TOR: the Yin and Yang of cellular nutrient sensing and growth control. Cell Metab 31, 472-492.
DOI PMID |
| [26] |
Greenham K, McClung CR (2015). Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16, 598-610.
DOI URL |
| [27] |
Heitman J, Movva NR, Hall MN (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905-909.
PMID |
| [28] |
Inaba JI, Nagy PD (2018). Tombusvirus RNA replication depends on the TOR pathway in yeast and plants. Virology 519, 207-222.
DOI URL |
| [29] |
Kahan BD (2003). Discoverer of the treasure from a barren island: suren sehgal (10 February 1932 to 21 January 2003). Transplantation 76, 623-624.
DOI URL |
| [30] |
Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, Blenis J (2013). Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol Cell 49, 172-185.
DOI URL |
| [31] |
Laplante M, Sabatini DM (2012). mTOR signaling in growth control and disease. Cell 149, 274-293.
DOI PMID |
| [32] |
Li B, Wang Y, Zhang YY, Tian WW, Chong K, Jang JC, Wang L (2019). PRR5, 7 and 9 positively modulate TOR signaling-mediated root cell proliferation by repressing TANDEM ZINC FINGER 1 in Arabidopsis. Nucleic Acids Res 47, 5001-5015.
DOI URL |
| [33] |
Li L, Liu KH, Sheen J (2021). Dynamic nutrient signaling networks in plants. Annu Rev Cell Dev Biol 37, 341-367.
DOI URL |
| [34] |
Li XJ, Cai WG, Liu YL, Li H, Fu LW, Liu ZY, Xu L, Liu HT, Xu TD, Xiong Y (2017). Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA 114, 2765-2770.
DOI URL |
| [35] | Liu GY, Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21, 183-203. |
| [36] |
Liu YL, Duan XL, Zhao XD, Ding WL, Wang YW, Xiong Y (2021). Diverse nitrogen signals activate convergent ROP2- TOR signaling in Arabidopsis. Dev Cell 56, 1283-1295.
DOI URL |
| [37] |
Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99, 6422-6427.
DOI URL |
| [38] |
Nukarinen E, Nägele T, Pedrotti L, Wurzinger B, Mair A, Landgraf R, Börnke F, Hanson J, Teige M, Baena-Gonzalez E, Dröge-Laser W, Weckwerth W (2016). Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci Rep 6, 31697.
DOI PMID |
| [39] |
O'Leary BM, Oh GGK, Lee CP, Millar AH (2020). Metabolite regulatory interactions control plant respiratory metabolism via target of rapamycin (TOR) kinase activation. Plant Cell 32, 666-682.
DOI URL |
| [40] |
Pacheco JM, Canal MV, Pereyra CM, Welchen E, Martínez-Noël GMA, Estevez JM (2021). The tip of the iceberg: emerging roles of TORC1, and its regulatory functions in plant cells. J Exp Bot 72, 4085-4101.
DOI PMID |
| [41] |
Pfeiffer A, Janocha D, Dong YH, Medzihradszky A, Schöne S, Daum G, Suzaki T, Forner J, Langenecker T, Rempel E, Schmid M, Wirtz M, Hell R, Lohmann JU (2016). Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. eLife 5, e17023.
DOI URL |
| [42] |
Pu YT, Luo XJ, Bassham DC (2017). TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front Plant Sci 8, 1204.
DOI URL |
| [43] |
Ren MZ, Qiu SQ, Venglat P, Xiang DQ, Feng L, Selvaraj G, Datla R (2011). Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol 155, 1367-1382.
DOI URL |
| [44] |
Riegler S, Servi L, Scarpin MR, Godoy Herz MA, Kubac-zka MG, Venhuizen P, Meyer C, Brunkard JO, Kalyna M, Barta A, Petrillo E (2021). Light regulates alternative splicing outcomes via the TOR kinase pathway. Cell Rep 36, 109676.
DOI PMID |
| [45] |
Ryabova LA, Robaglia C, Meyer C (2019). Target of ra-pamycin kinase: central regulatory hub for plant growth and metabolism. J Exp Bot 70, 2211-2216.
DOI PMID |
| [46] |
Sabatini DM (2017). Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc Natl Acad Sci USA 114, 11818-11825.
DOI URL |
| [47] |
Saxton RA, Sabatini DM (2017). mTOR signaling in growth, metabolism, and disease. Cell 168, 960-976.
DOI URL |
| [48] |
Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA (2013). TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J 32, 1087-1102.
DOI PMID |
| [49] |
Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA (2011). Viral factor TAV recruits TOR/S6K1 signaling to activate reinitiation after long ORF translation. EMBO J 30, 1343-1356.
DOI PMID |
| [50] |
Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang ZB, Chicher J, Hammann P, Ryabova LA (2017). GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J 36, 886-903.
DOI PMID |
| [51] |
Sehgal SN, Baker H, Vézina C (1975). Rapamycin (AY-22, 989), a new antifungal antibiotic. II. Fermentation, isola-tion and characterization. J Antibiot 28, 727-732.
PMID |
| [52] |
Sharma M, Banday ZZ, Shukla BN, Laxmi A (2019). Glu-cose-regulated HLP1 acts as a key molecule in governing thermomemory. Plant Physiol 180, 1081-1100.
DOI PMID |
| [53] |
Shi L, Wu Y, Sheen J (2018). TOR signaling in plants: con-servation and innovation. Development 145, dev160887.
DOI URL |
| [54] |
Shimobayashi M, Hall MN (2014). Making new contacts: the mTOR network in metabolism and signaling crosstalk. Nat Rev Mol Cell Biol 15, 155-162.
DOI URL |
| [55] |
Smailov B, Alybayev S, Smekenov I, Mursalimov A, Sa-parbaev M, Sarbassov D, Bissenbaev A (2020). Wheat germination is dependent on plant target of rapamycin signaling. Front Cell Dev Biol 8, 606685.
DOI URL |
| [56] |
Soprano AS, Smetana JHC, Benedetti CE (2018). Regula-tion of tRNA biogenesis in plants and its link to plant growth and response to pathogens. Biochim Biophys Acta Gene Regul Mech 1861, 344-353.
DOI URL |
| [57] |
Su WL, Bao Y, Yu XQ, Xia XL, Liu C, Yin WL (2020). Autophagy and its regulators in response to stress in plants. Int J Mol Sci 21, 8889.
DOI URL |
| [58] |
Tatebe H, Shiozaki K (2017). Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules 7, 77.
DOI URL |
| [59] |
Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F (2004). Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol 134, 1527-1535.
DOI URL |
| [60] |
Van Leene J, Han C, Gadeyne A, Eeckhout D, Matthijs C, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Van de Cotte B, Stes E, Van Bel M, Storme V, Impens F, Gevaert K, Vandepoele K, De Smet I, De Jaeger G (2019). Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat Plants 5, 316-327.
DOI URL |
| [61] |
Wang LJ, Li HH, Zhao CZ, Li SF, Kong LY, Wu WW, Kong WS, Liu Y, Wei YY, Zhu JK, Zhang HR (2017). The in-hibition of protein translation mediated by AtGCN1 is es-sential for cold tolerance in Arabidopsis thaliana. Plant Cell Environ 40, 56-68.
DOI URL |
| [62] |
Wang PC, Zhao Y, Li ZP, Hsu CC, Liu X, Fu LW, Hou YJ, Du YY, Xie SJ, Zhang CG, Gao JH, Cao MJ, Huang XS, Zhu YF, Tang K, Wang XG, Tao WA, Xiong Y, Zhu JK (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell 69, 100-112.
DOI URL |
| [63] |
Wang Y, Qin YM, Li B, Zhang YY, Wang L (2020). Attenuated TOR signaling lengthens circadian period in Arabi-dopsis. Plant Signal Behav 15, 1710935.
DOI PMID |
| [64] |
Wu Y, Shi L, Li L, Fu LW, Liu YL, Xiong Y, Sheen J (2019). Integration of nutrient, energy, light, and hormone signaling via TOR in plants. J Exp Bot 70, 2227-2238.
DOI URL |
| [65] |
Xiong Y, McCormack M, Li L, Hall Q, Xiang CB, Sheen J (2013). Glucose-TOR signaling reprograms the transcriptome and activates meristems. Nature 496, 181-186.
DOI URL |
| [66] |
Xiong Y, Sheen J (2012). Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem 287, 2836-2842.
DOI PMID |
| [67] |
Xiong Y, Sheen J (2014). The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol 164, 499-512.
DOI PMID |
| [68] |
Xiong Y, Sheen J (2015). Novel links in the plant TOR kinase signaling network. Curr Opin Plant Biol 28, 83-91.
DOI PMID |
| [69] |
Yang HR, Wang J, Liu MJ, Chen XZ, Huang M, Tan D, Dong MQ, Wong CCL, Wang JW, Xu YH, Wang HW (2016). 4.4 Å resolution Cryo-EM structure of human mTOR complex 1. Protein Cell 7, 878-887.
DOI URL |
| [70] |
Yu YD, Zhong ZC, Ma LY, Xiang CB, Xu P, Xiong Y (2021). Direct sulphate-TOR signaling controls transcrip-tional reprogramming for shoot apex activation in Arabi-dopsis. bioRxiv doi: 10.1101/2021.03.09.434511.
DOI |
| [71] |
Yuan XB, Xu P, Yu YD, Xiong Y (2020). Glucose-TOR signaling regulates PIN2 stability to orchestrate auxin gradient and cell expansion in Arabidopsis root. Proc Natl Acad Sci USA 117, 32223-32225.
DOI URL |
| [72] |
Zhang N, Meng YY, Li X, Zhou Y, Ma LY, Fu LW, Schwarzländer M, Liu HT, Xiong Y (2019). Metabolite-mediated TOR signaling regulates the circadian clock in Arabidop-sis. Proc Natl Acad Sci USA 116, 25395-25397.
DOI URL |
| [73] |
Zhang ZZ, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C, Sun Y, Wang WF, Wang ZY (2016). TOR signaling pro-motes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr Biol 26, 1854-1860.
DOI URL |
| [74] |
Zhu JK (2016). Abiotic stress signaling and responses in plants. Cell 167, 313-324.
DOI URL |
| [1] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| [2] | 欧阳子龙, 贾湘璐, 石景忠, 滕维超, 刘秀. 生长调节剂对低温胁迫及复温下红海榄幼苗光合特性的影响[J]. 植物生态学报, 2025, 49(4): 638-652. |
| [3] | 李梦琦, 苗灵凤, 李大东, 龙奕帆, 叶冰冰, 杨帆. 海南东寨港红树林植物细根功能性状对不同潮位沉积物养分变化的响应[J]. 植物生态学报, 2025, 49(4): 552-561. |
| [4] | 王鸿梅, 袁蔚, 薛芳, 张召聪, 刘坤, 陈四龙. 植物SWEET基因参与逆境胁迫响应及其调控机制[J]. 植物学报, 2025, 60(4): 1-0. |
| [5] | 谢涛, 章一帆, 刘云辉, 游慧玉, 夏季奔奔, 马蓉, 张春妮, 华学军. 植物线粒体铁硫簇合成系统及其调控的研究进展[J]. 植物学报, 2025, 60(4): 1-0. |
| [6] | 陆珍, 谢光杰, Qaisar KHAN, 覃英, 黄毓燕, 郭道君, 杨婷婷, 杨丽涛, 邢永秀, 李杨瑞, 王震. 伯克霍尔德菌通过改善生理适应性及调节铝响应基因的表达增强甘蔗对铝胁迫的耐受性[J]. 植物生态学报, 2025, 49(3): 475-487. |
| [7] | 刘旭鹏, 王敏, 韩守安, 朱学慧, 王艳蒙, 潘明启, 张雯. 植物器官脱落调控因素及分子机理研究进展[J]. 植物学报, 2025, 60(3): 472-482. |
| [8] | 田奥, 李苇洁, 曹洋, 贾真真, 曾松. 马缨杜鹃幼苗生长对土壤水分胁迫的响应及其生理机制[J]. 植物生态学报, 2025, 49(3): 488-501. |
| [9] | 徐田甜, 杨培建, 周晓茜, 曹怡, 陈艳红, 刘国元, 张健, 魏辉. 紫薇GolS家族基因的理化特性与表达特征[J]. 植物学报, 2025, 60(3): 393-406. |
| [10] | 熊良林, 梁国鲁, 郭启高, 景丹龙. 基因可变剪接调控植物响应非生物胁迫研究进展[J]. 植物学报, 2025, 60(3): 435-448. |
| [11] | 刘盈麟, 李春明, 王昊, 武长路, 贺强. 长江口滨海湿地水鸟对底栖微藻群落的营养级联效应[J]. 植物生态学报, 2025, 49(3): 367-378. |
| [12] | 范惠玲, 路妍, 金文海, 王慧, 彭小星, 武学霞, 刘玉皎. 基于根系表型性状的蚕豆耐盐碱性鉴定与综合评价(长英文摘要)[J]. 植物学报, 2025, 60(2): 204-217. |
| [13] | 樊蓓, 任敏, 王延峰, 党峰峰, 陈国梁, 程国亭, 杨金雨, 孙会茹. 番茄SlWRKY45转录因子在响应低温和干旱胁迫中的功能(长英文摘要)[J]. 植物学报, 2025, 60(2): 186-203. |
| [14] | 石雅琦, 刘海双, 柯瑾, 马清, 王锁民. 植物环核苷酸门控离子通道研究进展[J]. 植物学报, 2025, 60(2): 294-306. |
| [15] | 张舒欣, 贾紫璇, 方涛, 刘一凡, 赵微, 王荣, 昌海超, 罗芳丽, 朱耀军, 于飞海. 植物抗逆能力评价方法研究进展[J]. 生物多样性, 2025, 33(2): 24168-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||