

植物学报 ›› 2015, Vol. 50 ›› Issue (2): 171-179.DOI: 10.3724/SP.J.1259.2015.00171 cstr: 32102.14.SP.J.1259.2015.00171
伍自力1,2, 余孟瑶2, 陈露2, 魏静2, 王晓琴3, 胡勇2, 闫妍2, 万平2,*( )
)
收稿日期:2014-10-27
接受日期:2015-02-02
出版日期:2015-03-01
发布日期:2015-04-10
通讯作者:
万平
作者简介:? 共同第一作者
基金资助:Zili Wu1, 2, Mengyao Yu2, Lu Chen2, Jing Wei2, Xiaoqin Wang3, Yong Hu2, Yan Yan2, Ping Wan2, *
Received:2014-10-27
Accepted:2015-02-02
Online:2015-03-01
Published:2015-04-10
Contact:
Wan Ping
About author:? These authors contributed equally to this paper
摘要: 镉是植物非必需的微量重金属元素, 镉胁迫引起植物细胞的代谢紊乱, 甚至导致细胞死亡。为了探索苔藓植物对镉胁迫的应答机制, 采用高通量测序及生物信息学技术分析了藓类模式植物——小立碗藓(Physcomitrella patens)在镉胁迫下的基因表达特征。结果表明, 在镉胁迫下, 小立碗藓细胞骨架组织、微管运动、DNA修复系统、端粒维护、配子体形成与有性生殖以及与氮代谢等相关基因的表达具有明显的镉胁迫应答特征, 暗示了这些基因可能共同参与小立碗藓对镉胁迫的调控反应。该研究结果为阐明植物对镉胁迫的应答机制提供了新的线索。
伍自力, 余孟瑶, 陈露, 魏静, 王晓琴, 胡勇, 闫妍, 万平. 小立碗藓对重金属镉胁迫的应答特征. 植物学报, 2015, 50(2): 171-179.
Zili Wu, Mengyao Yu, Lu Chen, Jing Wei, Xiaoqin Wang, Yong Hu, Yan Yan, Ping Wan. Transcriptome Analysis of Physcomitrella patens Response to Cadmium Stress by Bayesian Network. Chinese Bulletin of Botany, 2015, 50(2): 171-179.
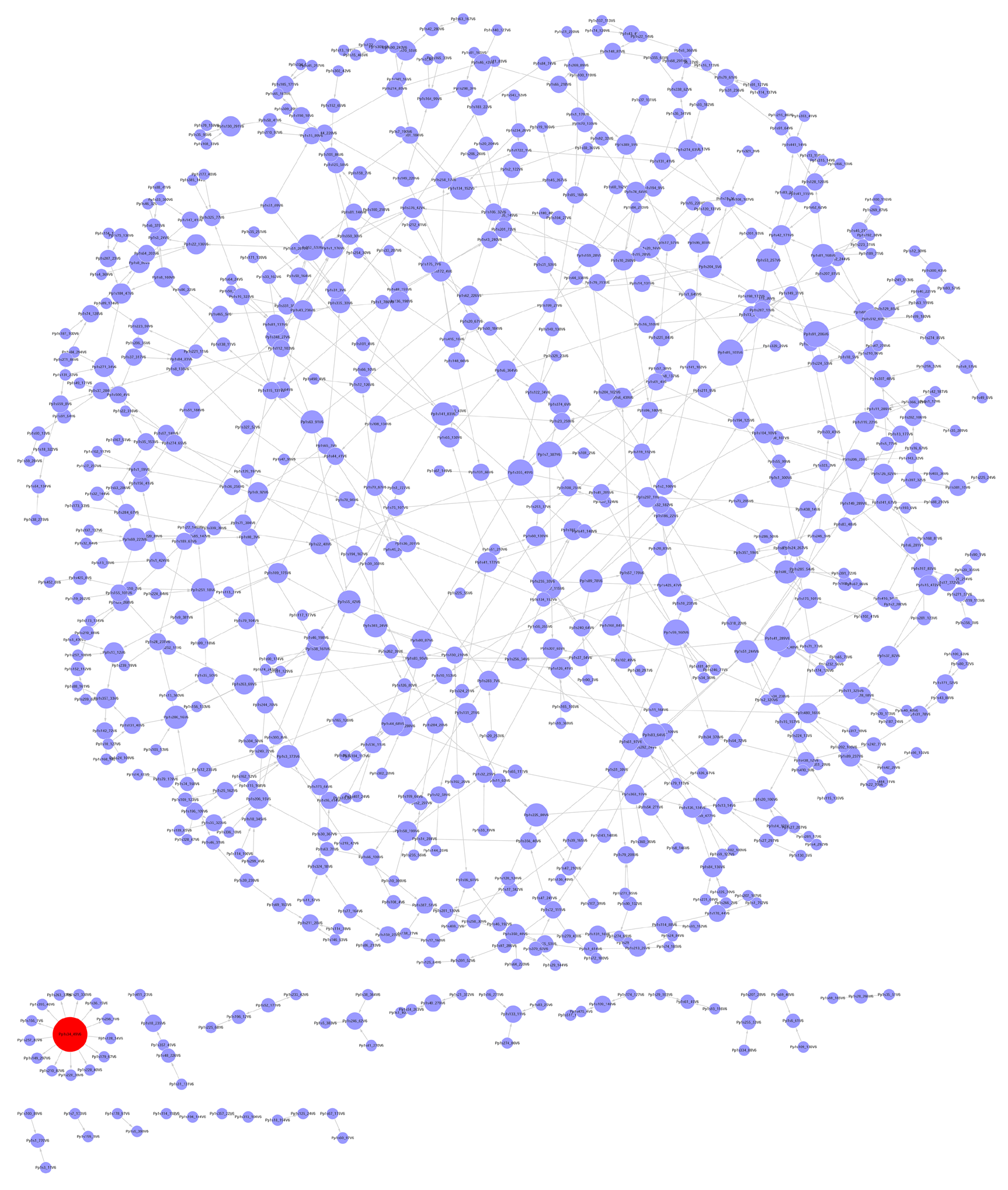
Figure 2 小立碗藓镉胁迫基因调控网络 该网络包含704个节点(基因), 788条边(调控关系)。图中红色节点的连接度为13
The gene regulatory network of Physcomitrella patens under cadmium stress The network consists of 704 nodes (genes) and 788 edges (regulation relation). The degree of the red node is thirteen
| Type | GO number | Biological processes | P value |
|---|---|---|---|
| 1 | GO:0007018 | Microtubule-based movement | 2.80E-06 |
| GO:0007017 | Microtubule-based process | 6.90E-05 | |
| GO:0007010 | Cytoskeleton organization | 0.018 8 | |
| 2 | GO:0006260 | DNA replication | 0.001 7 |
| GO:0006259 | DNA metabolic process | 0.002 5 | |
| GO:0006302 | Double-strand break repair | 0.015 3 | |
| GO:0000723 | Telomere maintenance | 0.033 4 | |
| GO:0032200 | Telomere organization | 0.033 4 | |
| 3 | GO:0007276 | Gamete generation | 0.033 4 |
| GO:0007292 | Female gamete generation | 0.033 4 | |
| GO:0019953 | Sexual reproduction | 0.033 4 | |
| 4 | GO:0019627 | Urea metabolic process | 0.033 4 |
| GO:0071941 | Nitrogen cycle metabolic process | 0.033 4 |
表1 小立碗藓镉胁迫基因调控网络中被富集的GO条目
Table 1 The enriched GO terms in Physcomitrella patens gene regulation network under cadmium stress
| Type | GO number | Biological processes | P value |
|---|---|---|---|
| 1 | GO:0007018 | Microtubule-based movement | 2.80E-06 |
| GO:0007017 | Microtubule-based process | 6.90E-05 | |
| GO:0007010 | Cytoskeleton organization | 0.018 8 | |
| 2 | GO:0006260 | DNA replication | 0.001 7 |
| GO:0006259 | DNA metabolic process | 0.002 5 | |
| GO:0006302 | Double-strand break repair | 0.015 3 | |
| GO:0000723 | Telomere maintenance | 0.033 4 | |
| GO:0032200 | Telomere organization | 0.033 4 | |
| 3 | GO:0007276 | Gamete generation | 0.033 4 |
| GO:0007292 | Female gamete generation | 0.033 4 | |
| GO:0019953 | Sexual reproduction | 0.033 4 | |
| 4 | GO:0019627 | Urea metabolic process | 0.033 4 |
| GO:0071941 | Nitrogen cycle metabolic process | 0.033 4 |
| Gene identification number | The homologous gene in Arabidopsis thaliana | Function |
|---|---|---|
| Pp1s34_49V6 | AT2G27290.1 | Protein of unknown function (DUF1279) |
| Pp1s352_53V6 | AT3G57060.2 | Chromosome condensation |
| Pp1s59_160V6 | AT3G44750.1 | Histone deacetylase 3 |
| Pp1s57_179V6 | AT5G18140.1 | Chaperone DnaJ-domain superfamily protein |
| Pp1s204_5V6 | AT5G20935.1 | Protein of unknown function |
| Pp1s68_291V6 | AT5G57590.1 | Adenosylmethionine-8-amino-7-oxononanoate transaminases |
| Pp1s13_454V6 | AT5G54910.1 | DEA (D/H)-box RNA helicase family protein |
| Pp1s133_35V6.1 | AT1G08260.1 | DNA polymerase epsilon catalytic subunit |
| Pp1s259_32V6.1 | AT1G79690.1 | Dipeptidyl-peptidase activity, hydrolase activity |
| Pp1s175_94V6.1 | AT4G01130.1 | GDSL-like Lipase/Acylhydrolase superfamily protein involved in lipid metabolic process |
表2 用于qPCR检测的10个基因
Table 2 A list of ten genes selected for qPCR analysis
| Gene identification number | The homologous gene in Arabidopsis thaliana | Function |
|---|---|---|
| Pp1s34_49V6 | AT2G27290.1 | Protein of unknown function (DUF1279) |
| Pp1s352_53V6 | AT3G57060.2 | Chromosome condensation |
| Pp1s59_160V6 | AT3G44750.1 | Histone deacetylase 3 |
| Pp1s57_179V6 | AT5G18140.1 | Chaperone DnaJ-domain superfamily protein |
| Pp1s204_5V6 | AT5G20935.1 | Protein of unknown function |
| Pp1s68_291V6 | AT5G57590.1 | Adenosylmethionine-8-amino-7-oxononanoate transaminases |
| Pp1s13_454V6 | AT5G54910.1 | DEA (D/H)-box RNA helicase family protein |
| Pp1s133_35V6.1 | AT1G08260.1 | DNA polymerase epsilon catalytic subunit |
| Pp1s259_32V6.1 | AT1G79690.1 | Dipeptidyl-peptidase activity, hydrolase activity |
| Pp1s175_94V6.1 | AT4G01130.1 | GDSL-like Lipase/Acylhydrolase superfamily protein involved in lipid metabolic process |
| 1 | Cabot C, Gallego B, Martos S, Barceló J, Poschenrieder C (2013). Signal cross talk in Arabidopsis exposed to cadmium, silicon, and Botrytis cinerea.Planta 237, 337-349. |
| 2 | Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, Shen YP, Zhang L, Deng XH, Luo JC, Deng XW, Chen ZL, Gu HY, Qu LJ (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family.Plant Mol Biol 60, 107-124. |
| 3 | Chmielowska-Bak J, Deckert J (2012). A common res- ponse to common danger? Comparison of animal and plant signaling pathways involved in cadmium sensing.J Cell Commun Signal 6, 191-204. |
| 4 | Chmielowska-Bak J, Deckert J (2013). Nitric oxide med- iates Cd-dependent induction of signaling-associated genes.Plant Signal Behav 8, e26664. |
| 5 | Chmielowska-Bak J, Gzyl J, Rucińska-Sobkowiak R, Arasimowicz-Jelonek M, Deckert J (2014). The new insights into cadmium sensing.Front Plant Sci 5, 245. |
| 6 | Corradi MG, Gorbi G, Ricci A, Torelli A, Bassi M (1995). Chromium-induced sexual reproduction gives rise to a Cr-tolerant progeny in Scenedesmus acutus.Ecotoxicol Environ Saf 32, 12-18. |
| 7 | DalCorso G, Farinati S, Furini A (2010). Regulatory networks of cadmium stress in plants.Plant Signal Behav 5, 663-667. |
| 8 | Dovgalyuk A, Kalynyak T, Blume YB (2003). Heavy metals have a different action from aluminium in disrupting microtubules in Allium cepa meristematic cells.Cell Biol Int 27, 193-195. |
| 9 | Ercal N, Gurer-Orhan H, Aykin-Burns N (2001). Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage.Curr Top Med Chem 1, 529-539. |
| 10 | Farinati S, DalCorso G, Varotto S, Furini A (2010). The Brassica juncea BjCdR15, an ortholog of Arabidopsis TGA3, is a regulator of cadmium uptake, transport and accumulation in shoots and confers cadmium tolerance in transgenic plants.New Phytol 185, 964-978. |
| 11 | Fojtová M, Fulnečková J, Fajkus J, Kovařík A (2002). Recovery of tobacco cells from cadmium stress is accompanied by DNA repair and increased telomerase activity.J Exp Bot 53, 2151-2158. |
| 12 | Hart JJ, Welch RM, Norvell WA, Sullivan LA, Kochian LV (1998). Characterization of cadmium binding, uptake, and translocation in intact seedlings of bread and durum wheat cultivars.Plant Physiol 116, 1413-1420. |
| 13 | Hartwig A, Schwerdtle T (2002). Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications.Toxicol Lett 127, 47-54. |
| 14 | Hepler PK, Hush JM (1996). Behavior of microtubules in living plant cells.Plant Physiol 112, 455-461. |
| 15 | Herbette S, Taconnat L, Hugouvieux V, Piette L, Magniette MLM, Cuine S, Auroy P, Richaud P, Forestier C, Bourguignon J, Renou JP, Vavasseur A, Leonhardt N (2006). Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88, 1751-1765. |
| 16 | Hsu YT, Kao CH (2003). Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings.Plant Cell Environ 26, 867-874. |
| 17 | Huang JJ, Okuka M, Lu WS, Tsibris JCM, McLean MP, Keefe DL, Liu L (2013). Telomere shortening and DNA damage of embryonic stem cells induced by cigarette smoke.Reprod Toxicol 35, 89-95. |
| 18 | Liu DH, Xue P, Meng QM, Zou J, Gu JG, Jiang WS (2009). Pb/Cu effects on the organization of microtubule cyto- skeleton in interphase and mitotic cells of Allium sativum L.Plant Cell Rep 28, 695-702. |
| 19 | Liu XM, Kim KE, Kim KC, Nguyen XC, Han HJ, Jung MS, Kim HS, Kim SH, Park HC, Yun DJ, Chung WS (2010). Cadmium activates Arabidopsis MPK3 and MPK6 via accumulation of reactive oxygen species.Phytochemistry 71, 614-618. |
| 20 | Ma WW, Xu WZ, Xu H, Chen YS, He ZY, Ma M (2010). Nitric oxide modulates cadmium influx during cadmium- induced programmed cell death in tobacco BY-2 cells.Planta 232, 325-335. |
| 21 | Nzengue Y, Steiman R, Garrel C, Lefèbvre E, Guiraud P (2008). Oxidative stress and DNA damage induced by cadmium in the human keratinocyte HaCaT cell line: role of glutathione in the resistance to cadmium.Toxicology 243, 193-206. |
| 22 | Oono Y, Yazawa T, Kawahara Y, Kanamori H, Kobayashi F, Sasaki H, Mori S, Wu J, Handa H, Itoh T, Matsumoto T (2014). Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice. PLoS One 9, e96946. |
| 23 | Přibyl P, Cepák V, Zachleder V (2008). Cytoskeletal alterations in interphase cells of the green alga Spirogyra decimina in response to heavy metals exposure: II. The effect of aluminium, nickel and copper.Toxicol In Vitro 22, 1160-1168. |
| 24 | Qi XT, Zhang YX, Chai TY (2007). Characterization of a novel plant promoter specifically induced by heavy metal and identification of the promoter regions conferring he- avy metal responsiveness.Plant Physiol 143, 50-59. |
| 25 | Rai V, Vajpayee P, Singh SN, Mehrotra S (2004). Effect of chromium accumulation on photosynthetic pigments, oxidative stress defense system, nitrate reduction, proline level and eugenol content of Ocimum tenuiflorum L.Plant Sci 167, 1159-1169. |
| 26 | Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin IT, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Cho SH, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van de Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants.Science 319, 64-69. |
| 27 | Roth U, von Roepenack-Lahaye E, Clemens S (2006). Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+.J Exp Bot 57, 4003-4013. |
| 28 | Rother M, Krauss GJ, Grass G, Wesenberg D (2006). Sulphate assimilation under Cd2+ stress in Physcomitrella patens—combined transcript, enzyme and metabolite profiling. Plant Cell Environ 29, 1801-1811. |
| 29 | Singh I, Shah K (2014). Evidences for structural basis of altered ascorbate peroxidase activity in cadmium- stressed rice plants exposed to jasmonate.Biometals 27, 247-263. |
| 30 | van de Mortel JE, Schat H, Moerland PD, Ver Loren van Themaat E, van der Ent S, Blankestijn H, Ghandilyan A, Tsiatsiani S, Aarts MG (2008). Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens.Plant Cell Environ 31, 301-324. |
| 31 | Wang YC, Gao CQ, Liang YN, Wang C, Yang CP, Liu GF (2010). A novel bZIP gene from Tamarix hispida mediates physiological responses to salt stress in tobacco plants.J Plant Physiol 167, 222-230. |
| 32 | Weber M, Trampczynska A, Clemens S (2006). Compara- tive transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd2+-hypertolerant facultative metallophyte Arabidopsis halleri.Plant Cell Environ 29, 950-963. |
| 33 | Xiong J, Fu G, Tao L, Zhu C (2010). Roles of nitric oxide in alleviating heavy metal toxicity in plants.Arch Biochem Biophys 497, 13-20. |
| 34 | Xiong J, Lu H, Lu KX, Duan YX, An LY, Zhu C (2009). Cadmium decreases crown root number by decreasing endogenous nitric oxide, which is indispensable for crown root primordia initiation in rice seedlings.Planta 230, 599-610. |
| 35 | Ye Y, Li Z, Xing D (2013). Nitric oxide promotes MPK6-mediated caspase-3-like activation in cadmium- induced Arabidopsis thaliana programmed cell death.Plant Cell Environ 36, 1-15. |
| 36 | Yourtchi MS, Bayat H (2013). Effect of cadmium toxicity on growth, cadmium accumulation and macronutrient content of durum wheat (Dena CV.).Int J Agri Crop Sci 6, 1099-1103. |
| [1] | 王鸿梅, 袁蔚, 薛芳, 张召聪, 刘坤, 陈四龙. 植物SWEET基因参与逆境胁迫响应及其调控机制[J]. 植物学报, 2025, 60(4): 1-0. |
| [2] | 熊良林, 梁国鲁, 郭启高, 景丹龙. 基因可变剪接调控植物响应非生物胁迫研究进展[J]. 植物学报, 2025, 60(3): 435-448. |
| [3] | 夏婧, 饶玉春, 曹丹芸, 王逸, 柳林昕, 徐雅婷, 牟望舒, 薛大伟. 水稻中乙烯生物合成关键酶OsACS和OsACO调控机制研究进展[J]. 植物学报, 2024, 59(2): 291-301. |
| [4] | 罗正明, 刘晋仙, 张变华, 周妍英, 郝爱华, 杨凯, 柴宝峰. 不同退化阶段亚高山草甸土壤原生生物群落多样性特征及驱动因素[J]. 生物多样性, 2023, 31(8): 23136-. |
| [5] | 张仲富, 王四海, 杨卫, 陈剑. 蒜头果根际细菌群落结构与功能特征对其健康状态的响应[J]. 植物生态学报, 2023, 47(7): 1020-1031. |
| [6] | 毛莹儿, 周秀梅, 王楠, 李秀秀, 尤育克, 白尚斌. 毛竹扩张对杉木林土壤细菌群落的影响[J]. 生物多样性, 2023, 31(6): 22659-. |
| [7] | 戴若惠, 钱心妤, 孙静蕾, 芦涛, 贾绮玮, 陆天麒, 路梅, 饶玉春. 水稻叶色调控机制及相关基因研究进展[J]. 植物学报, 2023, 58(5): 799-812. |
| [8] | 蔡淑钰, 刘建新, 王国夫, 吴丽元, 宋江平. 褪黑素促进镉胁迫下番茄种子萌发的调控机理[J]. 植物学报, 2023, 58(5): 720-732. |
| [9] | 白雪, 李玉靖, 景秀清, 赵晓东, 畅莎莎, 荆韬羽, 刘晋汝, 赵鹏宇. 谷子及其根际土壤微生物群落对铬胁迫的响应机制[J]. 植物生态学报, 2023, 47(3): 418-433. |
| [10] | 赵雯, 王丹丹, 热依拉·木民, 黄开钏, 刘顺, 崔宝凯. 阿尔山地区兴安落叶松林土壤微生物群落结构[J]. 生物多样性, 2023, 31(2): 22258-. |
| [11] | 夏凡, 杨婧, 李建, 史洋, 盖立新, 黄文华, 张经纬, 杨南, 高福利, 韩莹莹, 鲍伟东. 北京地区四个豹猫亚种群肠道菌群的组成[J]. 生物多样性, 2022, 30(9): 22103-. |
| [12] | 李聪, 齐立娟, 谷晓峰, 李继刚. 植物光信号途径重要新调控因子TZP的研究进展[J]. 植物学报, 2022, 57(5): 579-587. |
| [13] | 石水琴, 秦华光, 张静静, 韩钰, 余淏, 彭怡宁, 杨邵, 汪嘉怡, 何光宇, 岂泽华, 吴文杰, 朱星雨, 饶玉春, 穆丹. 濒危植物大别山五针松根际细菌群落特征与功能分析[J]. 植物学报, 2022, 57(4): 457-467. |
| [14] | 徐海霞, 何静, 易航, 王丽. 镉胁迫下地钱转录组的性别特异性响应机制[J]. 植物学报, 2022, 57(2): 182-196. |
| [15] | 孙翌昕, 李英滨, 李玉辉, 李冰, 杜晓芳, 李琪. 高通量测序技术在线虫多样性研究中的应用[J]. 生物多样性, 2022, 30(12): 22266-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||