

植物学报 ›› 2017, Vol. 52 ›› Issue (6): 723-732.DOI: 10.11983/CBB16237 cstr: 32102.14.CBB16237
宋慧芳, 刘海双, 杨义明, 范书田, 李昌禹*( ), 艾军*(
), 艾军*( )
)
收稿日期:2016-12-02
接受日期:2017-01-24
出版日期:2017-11-01
发布日期:2018-02-22
通讯作者:
李昌禹,艾军
基金资助:
Song Huifang, Liu Haishuang, Yang Yiming, Fan Shutian, Li Changyu*( ), Ai Jun*(
), Ai Jun*( )
)
Received:2016-12-02
Accepted:2017-01-24
Online:2017-11-01
Published:2018-02-22
Contact:
Li Changyu,Ai Jun
摘要: 对山葡萄(Vitis amurensis)种质资源样品的ITS、ITS2、psbA-trnH、rbcL和matK序列进行PCR扩增及测序, 优化PCR反应的退火温度, 比较各序列的扩增效率、测序成功率、品种间和品种内的差异及barcoding gap图, 使用BLAST和NJ树法比较不同序列的鉴定能力, 最终从5条DNA片段中筛选出可用于山葡萄种质资源鉴定的DNA条形码通用序列。结果表明, 在采集的11份33个山葡萄样品中, psbA-trnH和ITS2序列的扩增与测序成功率较高, 其品种间、品种内差异及barcoding gap较ITS、rbcL和matK序列具有明显的优势, 且ITS2序列能够鉴别psbA-trnH序列无法鉴别的品种。实验证明, ITS2和psbA-trnH序列是较适合鉴别山葡萄资源的DNA条形码序列组合。DNA条形码弥补了形态学鉴定的不足, 可为山葡萄种质资源的准确鉴定提供科学依据。
宋慧芳, 刘海双, 杨义明, 范书田, 李昌禹, 艾军. 山葡萄种质资源DNA条形码通用序列的筛选. 植物学报, 2017, 52(6): 723-732.
Song Huifang, Liu Haishuang, Yang Yiming, Fan Shutian, Li Changyu, Ai Jun. Screening of Universal DNA Barcodes for Vitis amurensis. Chinese Bulletin of Botany, 2017, 52(6): 723-732.
| Number | Varieties name | Locality of origin | Parents or source | Flower type |
|---|---|---|---|---|
| 1 | Zuoyouhong | Zuojia, Jilin | Varieties | Bisexual |
| 2 | Shuanghong | Zuojia, Jilin | Varieties | Bisexual |
| 3 | Zuoshan1 | Zuojia, Jilin | Wild resource | Male |
| 4 | Zuoshan2 | Zuojia, Jilin | Wild resource | Male |
| 5 | 4N1 | Zuojia, Jilin | Genetic material | Tetraploid |
| 6 | 4N2 | Zuojia, Jilin | Genetic material | Tetraploid |
| 7 | Shuangqing | Zuojia, Jilin | Varieties | Bisexual |
| 8 | Shuangfeng | Zuojia, Jilin | Varieties | Bisexual |
| 9 | Shuangyou | Ji’an, Jilin | Varieties | Bisexual |
| 10 | 75047 | Shangzhi, Heilongjiang | Wild resource | Female |
| 11 | 73061 | Dunhua, Jilin | Wild resource | Female |
表1 山葡萄品种信息
Table 1 Information of Vitis amurensis varieties
| Number | Varieties name | Locality of origin | Parents or source | Flower type |
|---|---|---|---|---|
| 1 | Zuoyouhong | Zuojia, Jilin | Varieties | Bisexual |
| 2 | Shuanghong | Zuojia, Jilin | Varieties | Bisexual |
| 3 | Zuoshan1 | Zuojia, Jilin | Wild resource | Male |
| 4 | Zuoshan2 | Zuojia, Jilin | Wild resource | Male |
| 5 | 4N1 | Zuojia, Jilin | Genetic material | Tetraploid |
| 6 | 4N2 | Zuojia, Jilin | Genetic material | Tetraploid |
| 7 | Shuangqing | Zuojia, Jilin | Varieties | Bisexual |
| 8 | Shuangfeng | Zuojia, Jilin | Varieties | Bisexual |
| 9 | Shuangyou | Ji’an, Jilin | Varieties | Bisexual |
| 10 | 75047 | Shangzhi, Heilongjiang | Wild resource | Female |
| 11 | 73061 | Dunhua, Jilin | Wild resource | Female |
| Fragment | Eight annealing temperature gradient (°C) | Annealing temperature (°C) |
|---|---|---|
| ITS2 | 54.8-55.4-56.0-56.6-57.2-57.8-58.4-59.0 | 56.0 |
| psbA-trnH | 52.2-52.8-53.4-54.0-54.6-55.2-55.8-56.4 | 55.2 |
| matK | 48.9-49.5-50.1-50.7-51.3-51.9-52.5-53.1 | 50.1 |
| rbcL | 52.1-52.7-53.3-53.9-54.5-55.1-55.7-56.3 | 54.5 |
| ITS | 49.9-50.5-51.1-51.7-52.3-52.9-53.5-54.1 | 52.9 |
表2 山葡萄DNA条形码标记引物序列及退火温度
Table 2 The primer information and annealing temperature of PCR from Vitis amurensis
| Fragment | Eight annealing temperature gradient (°C) | Annealing temperature (°C) |
|---|---|---|
| ITS2 | 54.8-55.4-56.0-56.6-57.2-57.8-58.4-59.0 | 56.0 |
| psbA-trnH | 52.2-52.8-53.4-54.0-54.6-55.2-55.8-56.4 | 55.2 |
| matK | 48.9-49.5-50.1-50.7-51.3-51.9-52.5-53.1 | 50.1 |
| rbcL | 52.1-52.7-53.3-53.9-54.5-55.1-55.7-56.3 | 54.5 |
| ITS | 49.9-50.5-51.1-51.7-52.3-52.9-53.5-54.1 | 52.9 |
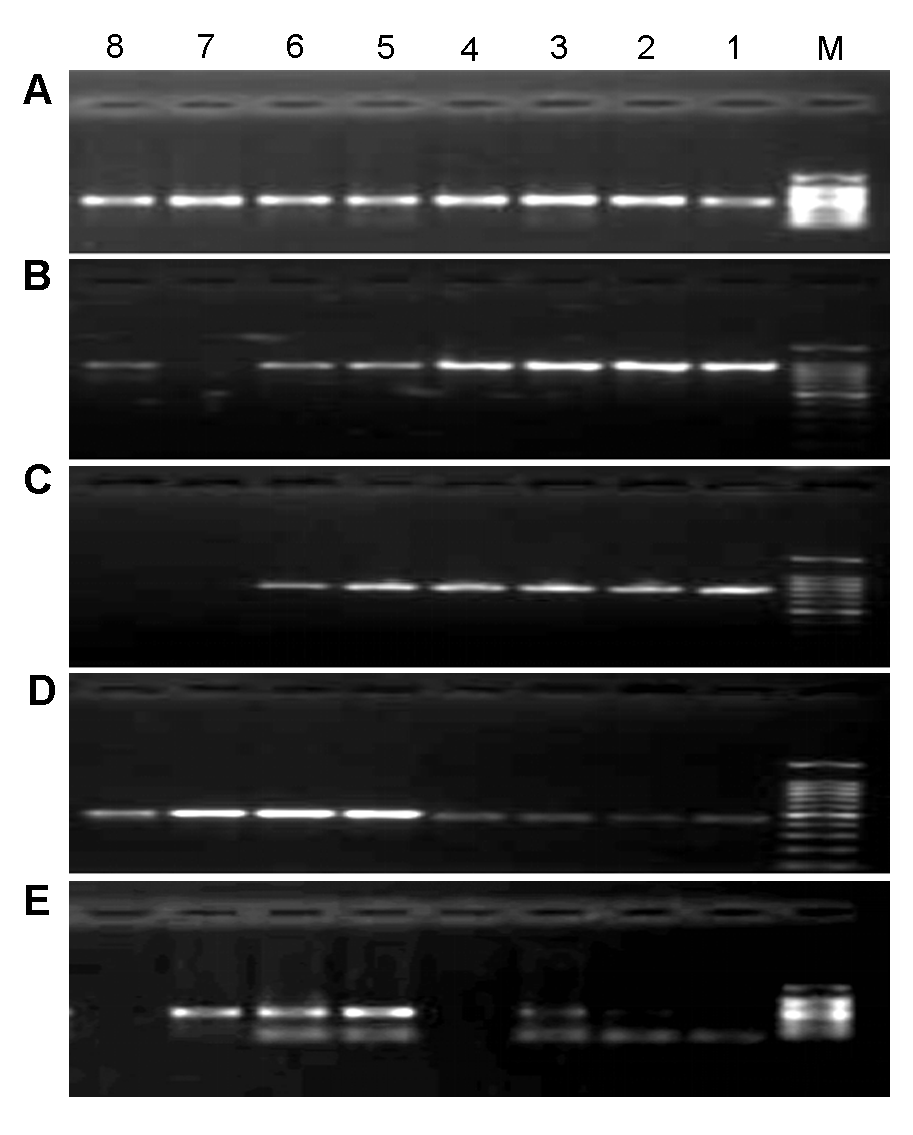
图1 5个候选序列PCR反应的梯度退火温度(A) ITS2; (B) matK; (C) rbcL; (D) psbA-trnH; (E) ITS。M: DNA marker DL2000; 1-8: 45-60°C之间的温度梯度
Figure 1 Gradient annealing temperature of PCR reaction of 5 candidate sequences(A) ITS2; (B) matK; (C) rbcL; (D) psbA-trnH; (E) ITS. M: DNA marker DL2000; 1-8: Gradient annealing temperature among 45-60°C
| Marker | Amplification efficiency (%) | Sequencing success rate (%) | Effective sequence ratio (%) |
|---|---|---|---|
| ITS2 | 100.0 | 96.9 | 96.9 |
| matK | 100.0 | 96.9 | 96.9 |
| psbA-trnH | 100.0 | 100.0 | 100.0 |
| rbcL | 96.9 | 90.9 | 88.1 |
| ITS | 45.5 | 30.3 | 13.8 |
表3 不同DNA条形码序列扩增获得的有效序列比例
Table 3 The effective sequence ratio obtained by PCR am- plification of five DNA barcode sequences
| Marker | Amplification efficiency (%) | Sequencing success rate (%) | Effective sequence ratio (%) |
|---|---|---|---|
| ITS2 | 100.0 | 96.9 | 96.9 |
| matK | 100.0 | 96.9 | 96.9 |
| psbA-trnH | 100.0 | 100.0 | 100.0 |
| rbcL | 96.9 | 90.9 | 88.1 |
| ITS | 45.5 | 30.3 | 13.8 |
| Potential barcode | Aligned length (bp) | Number of variable sites | Mean intra- distance | Mean inter- distance | Average of GC content (%) |
|---|---|---|---|---|---|
| ITS2 | 483 | 397 | 0.0015 | 0.1162 | 64.50 |
| matK | 896 | 44 | 0.0032 | 0.0110 | 35.40 |
| psbA-trnH | 422 | 238 | 0.0089 | 0.0921 | 27.50 |
| rbcL | 697 | 82 | 0.0068 | 0.0180 | 44.20 |
| ITS2+matK | 1379 | 441 | 0.0024 | 0.0652 | 49.95 |
| ITS2+psbA-trnH | 905 | 635 | 0.0058 | 0.0985 | 46.00 |
| ITS2+rbcL | 1180 | 479 | 0.0045 | 0.0655 | 54.35 |
| matK+psbA-trnH | 1318 | 282 | 0.0064 | 0.0550 | 31.45 |
| matK+rbcL | 1543 | 126 | 0.0051 | 0.0220 | 41.35 |
| psbA-trnH+rbcL | 1119 | 320 | 0.0076 | 0.0560 | 39.80 |
表4 4个候选序列/序列组合的长度、GC含量及品种间和品种内差异
Table 4 Measures of inter-varieties and intra-varieties divergence locus length and average of GC content for 4 candidate barcodes/combination sequences
| Potential barcode | Aligned length (bp) | Number of variable sites | Mean intra- distance | Mean inter- distance | Average of GC content (%) |
|---|---|---|---|---|---|
| ITS2 | 483 | 397 | 0.0015 | 0.1162 | 64.50 |
| matK | 896 | 44 | 0.0032 | 0.0110 | 35.40 |
| psbA-trnH | 422 | 238 | 0.0089 | 0.0921 | 27.50 |
| rbcL | 697 | 82 | 0.0068 | 0.0180 | 44.20 |
| ITS2+matK | 1379 | 441 | 0.0024 | 0.0652 | 49.95 |
| ITS2+psbA-trnH | 905 | 635 | 0.0058 | 0.0985 | 46.00 |
| ITS2+rbcL | 1180 | 479 | 0.0045 | 0.0655 | 54.35 |
| matK+psbA-trnH | 1318 | 282 | 0.0064 | 0.0550 | 31.45 |
| matK+rbcL | 1543 | 126 | 0.0051 | 0.0220 | 41.35 |
| psbA-trnH+rbcL | 1119 | 320 | 0.0076 | 0.0560 | 39.80 |
| w+ | w- | Inter relative ranks | n | P value | Result |
|---|---|---|---|---|---|
| ITS2 | rbcL | w+=33035.00, w-=13630.00 | 319 | 0.000 | P<0.01, ITS2>rbcL |
| ITS2 | matK | w+=1444453.00, w-=0.00 | 577 | 0.000 | P<0.01, ITS2>matK |
| ITS2 | psbA-trnH | w+=11562.00, w-=8660.00 | 500 | 0.000 | P<0.01, ITS2>psbA-trnH |
| rbcL | matK | w+=8673.00, w-=6903.00 | 319 | 0.174 | P>0.05, rbcL=matK |
| rbcL | psbA-trnH | w+=4350.00, w-=8720.00 | 620 | 0.000 | P<0.01, rbcL<psbA-trnH |
| matK | psbA-trnH | w+=12556.00, w-=25330.00 | 422 | 0.000 | P<0.01, matK<psbA-trnH |
表5 候选序列品种间差异的Wilcoxon检验
Table 5 Wilcoxon signes tests for inter-varieties divergences of candidate sequences
| w+ | w- | Inter relative ranks | n | P value | Result |
|---|---|---|---|---|---|
| ITS2 | rbcL | w+=33035.00, w-=13630.00 | 319 | 0.000 | P<0.01, ITS2>rbcL |
| ITS2 | matK | w+=1444453.00, w-=0.00 | 577 | 0.000 | P<0.01, ITS2>matK |
| ITS2 | psbA-trnH | w+=11562.00, w-=8660.00 | 500 | 0.000 | P<0.01, ITS2>psbA-trnH |
| rbcL | matK | w+=8673.00, w-=6903.00 | 319 | 0.174 | P>0.05, rbcL=matK |
| rbcL | psbA-trnH | w+=4350.00, w-=8720.00 | 620 | 0.000 | P<0.01, rbcL<psbA-trnH |
| matK | psbA-trnH | w+=12556.00, w-=25330.00 | 422 | 0.000 | P<0.01, matK<psbA-trnH |
| w+ | w- | Inter relative ranks | n | P value | Result |
|---|---|---|---|---|---|
| ITS2 | rbcL | w+=39345.00, w-=18966.00 | 384 | 0.000 | P<0.01, ITS2>rbcL |
| ITS2 | matK | w+=142845.00, w-=0.00 | 577 | 0.000 | P<0.01, ITS2>matK |
| ITS2 | psbA-trnH | w+=17550.00, w-=18326.00 | 366 | 0.056 | P>0.05, ITS2=psbA-trnH |
| rbcL | matK | w+=11952.00, w-=5253.00 | 384 | 0.000 | P<0.01, rbcL>matK |
| rbcL | psbA-trnH | w+=5968.00, w-=13589.00 | 469 | 0.000 | P<0.01, rbc<psbA-trnH |
| matK | psbA-trnH | w+=3985.00, w-=12578.00 | 580 | 0.000 | P<0.01, matK<psbA-trnH |
表6 候选序列品种内差异的Wilcoxon检验
Table 6 Wilcoxon signes tests for intra-varieties divergences of candidate sequences
| w+ | w- | Inter relative ranks | n | P value | Result |
|---|---|---|---|---|---|
| ITS2 | rbcL | w+=39345.00, w-=18966.00 | 384 | 0.000 | P<0.01, ITS2>rbcL |
| ITS2 | matK | w+=142845.00, w-=0.00 | 577 | 0.000 | P<0.01, ITS2>matK |
| ITS2 | psbA-trnH | w+=17550.00, w-=18326.00 | 366 | 0.056 | P>0.05, ITS2=psbA-trnH |
| rbcL | matK | w+=11952.00, w-=5253.00 | 384 | 0.000 | P<0.01, rbcL>matK |
| rbcL | psbA-trnH | w+=5968.00, w-=13589.00 | 469 | 0.000 | P<0.01, rbc<psbA-trnH |
| matK | psbA-trnH | w+=3985.00, w-=12578.00 | 580 | 0.000 | P<0.01, matK<psbA-trnH |
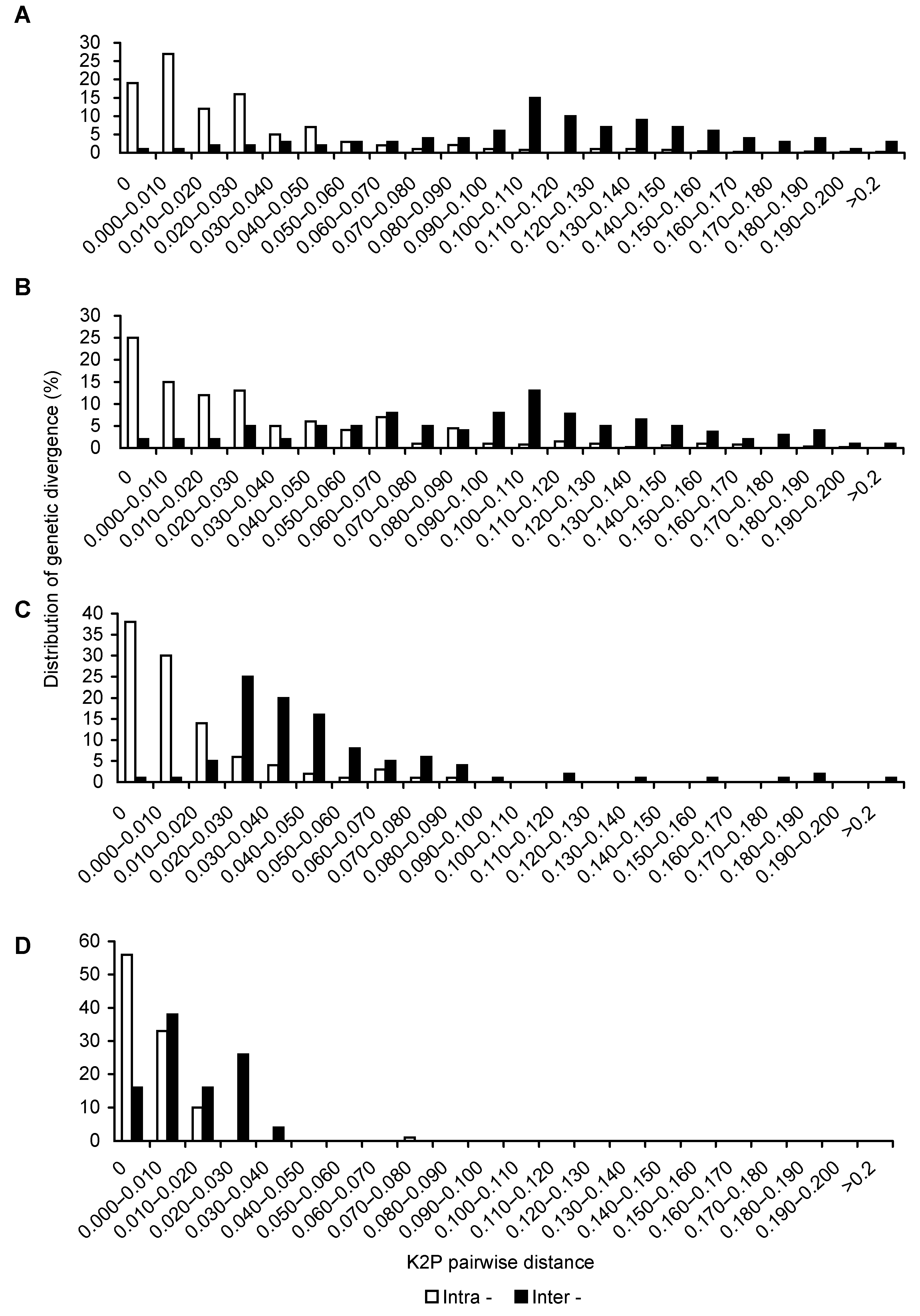
图2 山葡萄品种间和品种内变异barcoding gap图(A) ITS2序列; (B) psbA-trnH序列; (C) rbcL序列; (D) matK序列
Figure 2 Distribution for intra- and inter-varieties variation of Vitis amurensis(A) ITS2 sequence; (B) psbA-trnH sequence; (C) rbcL sequence; (D) matK sequence
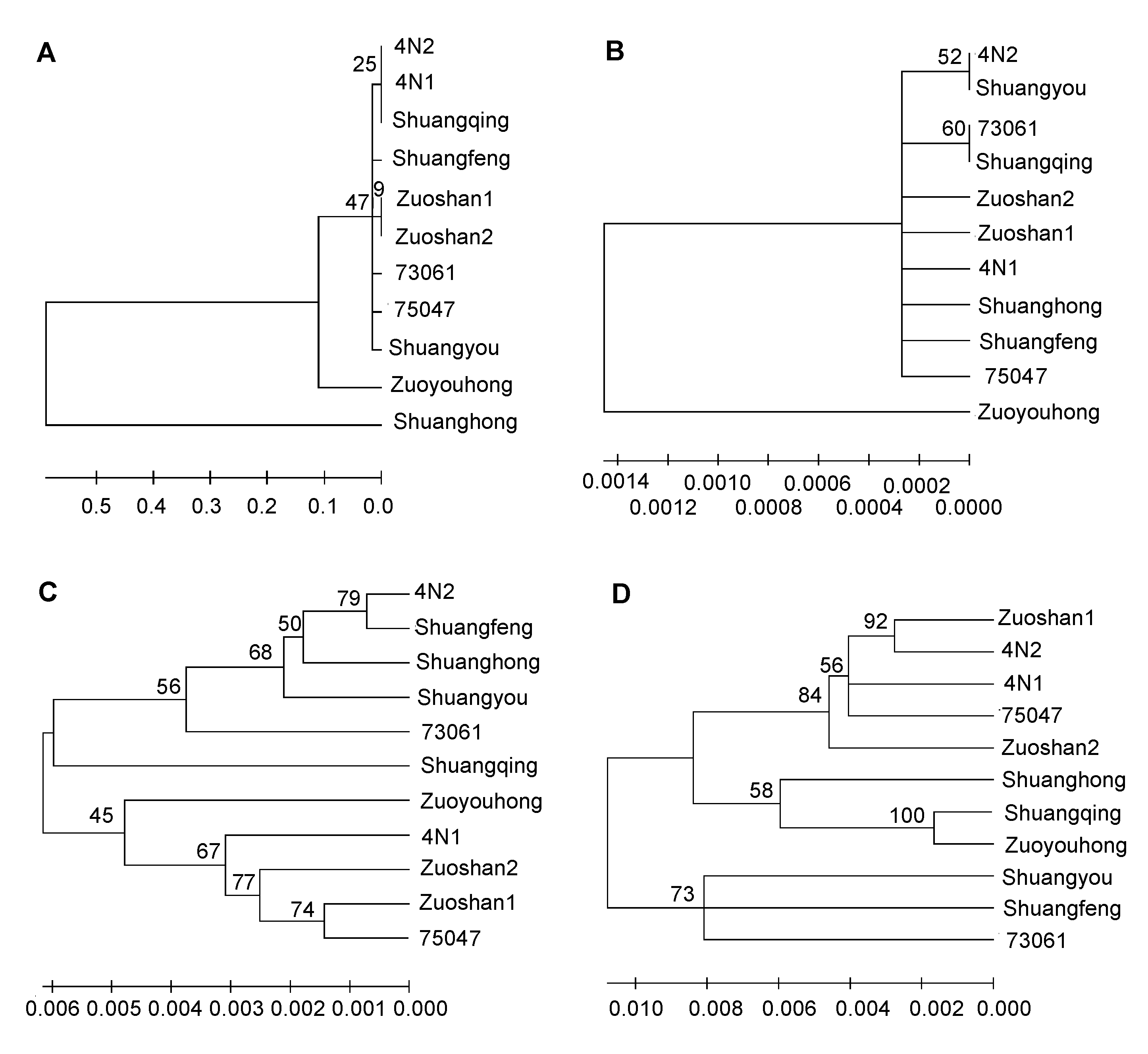
图3 利用不同序列构建的11份山葡萄资源品种系统进化树(A) ITS2序列; (B) psbA-trnH序列; (C) rbcL序列; (D) matK序列
Figure 3 Neighbor-joining (NJ) tree for 11 varieties of Vitis amurensis by using different sequences(A) ITS2 sequence; (B) psbA-trnH sequence; (C) rbcL sequence; (D) matK sequence
| [1] |
陈士林, 姚辉, 韩建萍, 辛天怡, 庞晓慧, 石林春, 罗焜, 宋经元, 侯典云, 石上梅, 钱忠直 (2013). 中药材DNA条形码分子鉴定指导原则. 中国中药杂志 38, 141-148.
DOI URL |
| [2] |
东秀珠, 沈德龙, 辛玉华 (2000). 16S rDNA同源性所揭示的双歧杆菌与有关细菌的亲缘关系. 生物多样性 8, 146-152.
DOI URL |
| [3] |
高健, 孟婉姮, 杜芳, 李俊清 (2015). 鸡爪槭种下分类群的DNA条形码筛选. 植物科学学报33, 734-743.
DOI URL |
| [4] |
李晓艳, 杨义明, 范书田, 王振兴, 艾军, 沈育杰 (2014). 山葡萄种质资源收集、保存、评价与利用研究进展. 河北林业科技 (5-6), 115-121.
DOI URL |
| [5] |
刘震, 陈科力, 罗焜, 潘宏林, 陈士林 (2010). 忍冬科药用植物DNA条形码通用序列的筛选. 中国中药杂志 35, 2527-2532.
DOI URL |
| [6] | 任保青, 陈之端 (2010). 植物DNA条形码技术. 植物学报45, 1-12. |
| [7] |
任阳阳, 张梦婷, 张嘉丽, 樊佳佳, 张晓存, 王俊, 刘霞 (2016). 虾脊兰属植物DNA条形码的确立. 世界中医药 11, 2425-2429.
DOI URL |
| [8] |
沈育杰, 赵淑兰, 杨义明, 李晓红, 宋润刚, 路文鹏 (2006). 我国山葡萄种质资源研究与利用现状. 特产研究28(3), 53-57.
DOI URL |
| [9] |
石志刚, 万如, 李彦龙, 王亚军, 马婷慧 (2016). 宁夏枸杞主要品种psbA-trnH的DNA条形码鉴定的初步研究. 农业科技与装备(6), 1-2, 7.
DOI URL |
| [10] |
宋润刚, 艾军, 李晓红, 杨义明, 沈育杰 (2009). 中国山葡萄产业的发展及对策. 中外葡萄与葡萄酒 (11), 64-69.
DOI URL |
| [11] |
王柯, 陈科力, 刘震, 陈士林 (2011). 锦葵科植物DNA条形码通用序列的筛选. 植物学报 46, 276-284.
DOI URL |
| [12] | 辛天怡, 姚辉, 罗焜, 向丽, 马晓冲, 韩建萍, 林余霖, 宋经元, 陈士林 (2012). 羌活药材ITS/ITS2条形码鉴定及其稳定性与准确性研究. 药学学报 47, 1098-1105. |
| [13] |
CBOL Plant Working Group (2009). A DNA barcode for land plants.Proc Natl Acad Sci USA 106, 12794-12797.
DOI URL PMID |
| [14] |
Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madri?án S, Petersen G, Seberg O, J?rgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough TG, Kelly L, Wilkinson M (2007). A proposal for a standardised protocol to barcode all land plants.Taxon 56, 295-299.
DOI URL |
| [15] |
Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species.PLoS One 5, e8613.
DOI URL PMID |
| [16] |
China Plant BOL Group, Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB, Fu CX, Zeng CX, Yan HF, Zhu YJ, Sun YS, Chen SY, Zhao L, Wang K, Yang T, Duan GW (2011). Comparative analysis of a large dataset indicates that internal trans- cribed spacer (ITS) should be incorporated into the core barcode for seed plants.Proc Natl Acad Sci USA 108, 19641-19646.
DOI URL PMID |
| [17] | Dassanayake RS, Gunawardene YINS, De Silva BDDNK (2008). ITS-2 secondary structures and phylogeny of Ano- pheles culicifacies species. Bioinformation 2, 456-460. |
| [18] |
Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, Percy DM, Hajibabaei M, Barrett SCH (2008). Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well.PLoS One 3, e2802.
DOI URL |
| [19] |
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003). Biological identifications through DNA barcodes.Proc Roy Soc B Biol Sci 270, 313-321.
DOI URL |
| [20] |
Hollingsworth PM (2008). DNA barcoding plants in biodiversity hot spots: progress and outstanding questions.Here- dity 101, 1-2.
DOI URL PMID |
| [21] | Kress WJ, Erickson DL (2007). A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One 2, e508. |
| [22] |
Keller A, Schleicher T, Schultz J, Müller T, Dandekar T, Wolf M (2009). 5.8S-28S rRNA interaction and HMM-ba- sed ITS2 annotation.Gene 430, 50-57.
DOI URL PMID |
| [23] |
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005). Use of DNA barcodes to identify flowering plants.Proc Natl Acad Sci USA 102, 8369-8374.
DOI URL PMID |
| [24] |
Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008). DNA barcoding the floras of bio- diversity hotspots.Proc Natl Acad Sci USA 105, 2923-2928.
DOI URL PMID |
| [25] |
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007). Clustal W and Clustal X version 2.0.Bioinformatics 23, 2947-2948.
DOI URL PMID |
| [26] |
Meyer CP, Paulay G (2005). DNA barcoding: error rates based on comprehensive sampling.PLoS Biol 3, e422.
DOI URL |
| [27] |
Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J (2008). Testing candidate plant barcode regions in the Myris- ticaceae.Mol Ecol Res 8, 480-490.
DOI URL PMID |
| [28] |
Ross HA, Murugan S, Li WLS (2008). Testing the reliability of genetic methods of species identification via simulation.Syst Biol 57, 216-230.
DOI URL PMID |
| [29] |
Selig C, Wolf M, Müller T, Dandekar T, Schultz J (2008). The ITS2 Database II: homology modelling RNA structure for molecular systematics.Nucleic Acids Res 36, D377-D380.
DOI URL PMID |
| [30] |
Slabbinck B, Dawyndt P, Martens M, De Vos P, De Baets B (2008). Taxon gap: a visualization tool for intra- and inter- species variation among individual biomarkers.Bioinformatics 24, 866-867.
DOI URL PMID |
| [31] |
Song JY, Yao H, Li Y, Li XW, Lin YL, Liu C, Han JP, Xie CX, Chen SL (2009). Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique.J Ethnopharmacol 124, 434-439.
DOI URL PMID |
| [32] |
Vinitha MR, Kumar US, Aishwarya K, Sabu M, Thomas G (2014). Prospects for discriminating Zingiberaceae species in India using DNA barcodes.J Integr Plant Biol 56, 760-773.
DOI URL PMID |
| [33] |
Yao H, Song JY, Ma XY, Liu C, Li Y, Xu HX, Han JP, Duan LS, Chen SL (2009). Identification of Dendrobium species by a candidate DNA barcode sequence: the chloroplast psbA-trnH intergenic region. Planta Med 75, 667-669.
DOI URL PMID |
| [1] | 陈龙 郭柯 勾晓华 赵秀海 马泓若. 祁连圆柏林群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 0-0. |
| [2] | 张琨, 钱敏, 汪阳, 李志华, 孔令娜, 李明洋, 马瑾煜, 努尔艾合麦提•玉苏普, 陈乙一, 成沂芮, 张焕仕, 覃凤飞, 渠晖. 紫花苜蓿耐阴性综合评价及其鉴定指标的筛选[J]. 植物生态学报, 2025, 49(5): 773-787. |
| [3] | 郭政, 邵香君, 鲁海雯, 侯丹, 孔思梦, 李翔宇, 刘华倩, 林新春. 马来甜龙竹多倍体高效诱导及鉴定[J]. 植物学报, 2025, 60(2): 246-255. |
| [4] | 范惠玲, 路妍, 金文海, 王慧, 彭小星, 武学霞, 刘玉皎. 基于根系表型性状的蚕豆耐盐碱性鉴定与综合评价(长英文摘要)[J]. 植物学报, 2025, 60(2): 204-217. |
| [5] | 巴苏艳, 赵春艳, 刘媛, 方强. 通过虫体花粉识别构建植物‒传粉者网络: 人工模型与AI模型高度一致[J]. 生物多样性, 2024, 32(6): 24088-. |
| [6] | 索南吉, 李博文, 吕汪汪, 王文颖, 拉本, 陆徐伟, 宋扎磋, 陈程浩, 苗琪, 孙芳慧, 汪诗平. 增温增水情景下钉柱委陵菜物候序列的变化及其抗冻性[J]. 植物生态学报, 2024, 48(2): 158-170. |
| [7] | 段政勇, 丁敏, 王宇卓, 丁艺冰, 陈凌, 王瑞云, 乔治军. 糜子SBP基因家族全基因组鉴定及表达分析[J]. 植物学报, 2024, 59(2): 231-244. |
| [8] | 罗小燕, 李强, 黄晓磊. 戴云山国家级自然保护区访花昆虫DNA条形码数据集[J]. 生物多样性, 2023, 31(8): 23236-. |
| [9] | 邢超, 林依, 周智强, 赵联军, 蒋仕伟, 林蓁蓁, 徐基良, 詹祥江. 基于DNA条形码技术构建王朗国家级自然保护区陆生脊椎动物遗传资源数据库及物种鉴定[J]. 生物多样性, 2023, 31(7): 22661-. |
| [10] | 王露露, 杨智, 杨永. 利用标本组学推进植物超级DNA条形码研究[J]. 植物学报, 2023, 58(5): 831-842. |
| [11] | 吴帆, 刘深云, 江虎强, 王茜, 陈开威, 李红亮. 中华蜜蜂和意大利蜜蜂秋冬期传粉植物多样性比较[J]. 生物多样性, 2023, 31(5): 22528-. |
| [12] | 孙蓉, 杨宇琭, 李亚军, 张会, 李旭凯. 谷子PLATZ转录因子基因家族的鉴定和分析[J]. 植物学报, 2023, 58(4): 548-559. |
| [13] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [14] | 白雪, 李玉靖, 景秀清, 赵晓东, 畅莎莎, 荆韬羽, 刘晋汝, 赵鹏宇. 谷子及其根际土壤微生物群落对铬胁迫的响应机制[J]. 植物生态学报, 2023, 47(3): 418-433. |
| [15] | 林春惠, 顾惠怡, 叶钦良, 张志坚, 钟智明, 易绮斐. 珍稀濒危植物大苞山茶种群结构与动态特征[J]. 植物生态学报, 2023, 47(12): 1684-1692. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||