

植物学报 ›› 2023, Vol. 58 ›› Issue (5): 831-842.DOI: 10.11983/CBB22194 cstr: 32102.14.CBB22194
• 专题论坛 • 上一篇
王露露1,2,3, 杨智1,2,3, 杨永1,2,3,*( )
)
收稿日期:2022-08-12
接受日期:2023-02-09
出版日期:2023-09-01
发布日期:2023-09-21
通讯作者:
*E-mail: yangyong@njfu.edu.cn
基金资助:
Wang Lulu1,2,3, Yang Zhi1,2,3, Yang Yong1,2,3,*( )
)
Received:2022-08-12
Accepted:2023-02-09
Online:2023-09-01
Published:2023-09-21
Contact:
*E-mail: yangyong@njfu.edu.cn
摘要: 物种的准确鉴定是生物资源利用和物种保护的重要基础。传统的物种鉴定依赖于分类学专业人员, 而培养分类学专业人员需要耗费大量时间, 是分类学知识利用的瓶颈。利用多通道电子检索表和智能识别也因个体发育阶段的不同和形态变异而难以准确鉴定物种。DNA条形码是基于DNA序列的鉴定技术, 不受个体发育和形态多态性的影响, 为物种快速鉴定提供了有效的解决方案。该文综述了DNA条形码的类型和发展趋势。DNA条形码的发展历史可分为2个阶段: (1) 以一代测序为序列获取方式的单片段或少量片段时期; (2) 以新一代高通量测序为序列获取方式的超级DNA条形码时期。该文比较了2个发展阶段DNA条形码的主要特点、研究方法和物种鉴定能力, 认为基于质体基因组的超级DNA条形码物种鉴定能力更强, 发展超级DNA条形码将成为植物条形码研究的主要趋势。同时进一步总结了超级DNA条形码发展所面临的困境和解决思路, 指出研究材料仍是当前限制超级DNA条形码发展的关键因素, 提出利用标本组学方法捕获大量历史馆藏标本的基因组序列, 可为开展超级DNA条形码研究提供省时、高效和经济的解决方案。通过挖掘植物基因组数据获得单拷贝和低拷贝核基因并应用于物种条形码研究, 不仅能提升分辨率, 而且可解决杂交和不完全谱系分选等复杂进化历史类群的分类问题。建议利用标本馆保存的珍贵标本资源建立植物超级DNA条形码参考序列数据库。
王露露, 杨智, 杨永. 利用标本组学推进植物超级DNA条形码研究. 植物学报, 2023, 58(5): 831-842.
Wang Lulu, Yang Zhi, Yang Yong. Plant Ultra-barcoding Using Herbariomics. Chinese Bulletin of Botany, 2023, 58(5): 831-842.
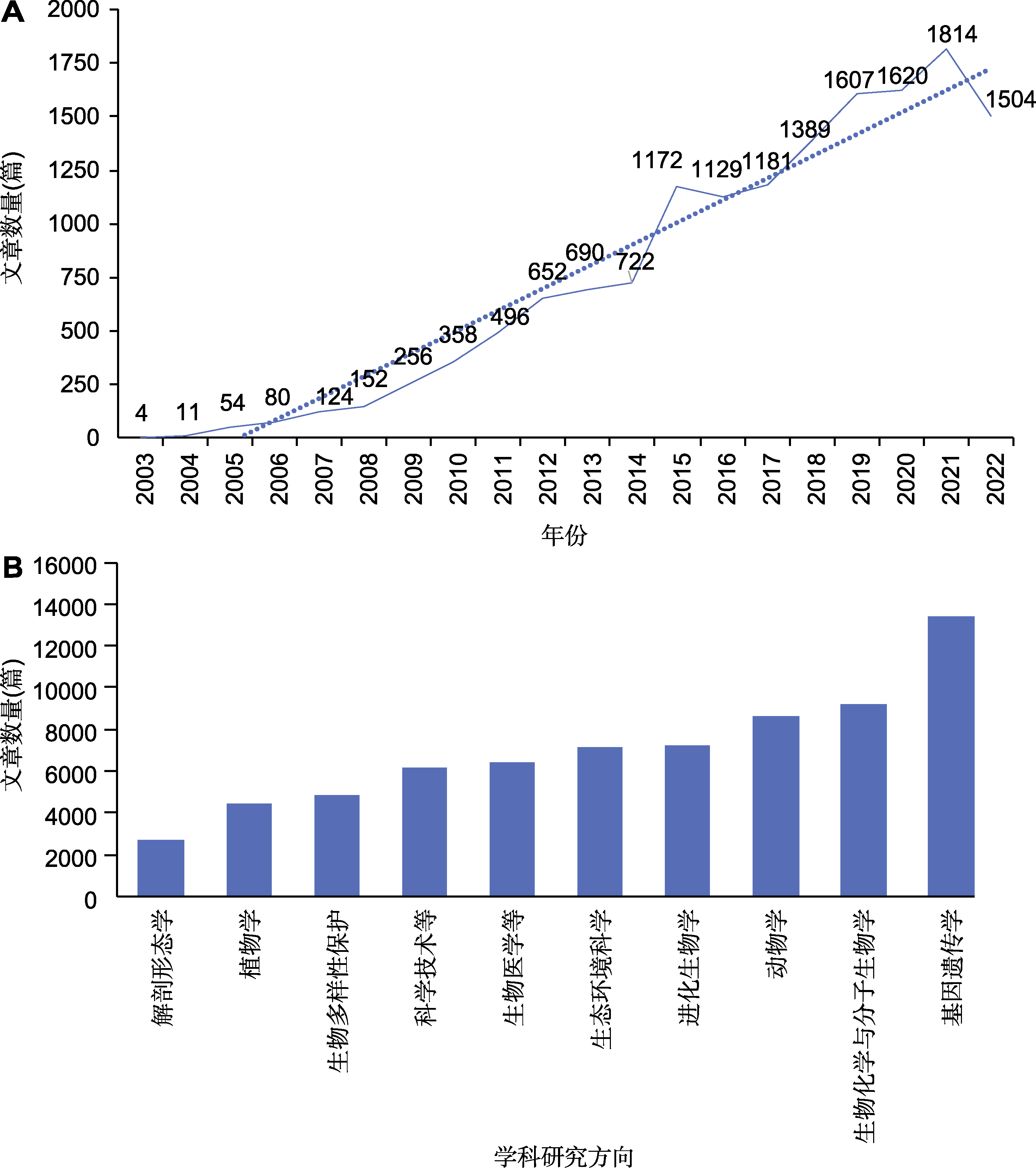
图1 Web of Science数据库历年发表的DNA条形码论文统计 (A) 2003-2022年DNA条形码相关文章数量; (B) 不同学科研究方向DNA条形码相关文章数量
Figure 1 Publications of DNA barcodes in the Web of Science database over years (A) Number of articles related to DNA barcoding published in 2003-2022; (B) Number of articles related to DNA barcoding published in different disciplines
| 测序技术 | 实验成本 | 最终成本 | DNA质量要求 | 同源序列的识别能力 | 生物信息学专门知识 | 是否适用于标本材料 |
|---|---|---|---|---|---|---|
| 简化基因组测序 | 高 | 低 | 中-高 | 难 | 中级/高级 | 否 |
| 转录组测序 | 低 | 低 | 中-高 | 相对容易 | 中级 | 否 |
| 杂交捕获测序 | 低 | 中 | 低-中 | 容易 | 中级/高级 | 是 |
| 扩增子测序 | 低 | 中 | 低-中 | 容易 | 初级 | 是 |
| 基因组浅层测序 | 低 | 中 | 低-中 | 容易 | 初级 | 是 |
表1 5种常用基因组划分方法的比较
Table 1 Comparison of five commonly used methods for genome partitioning
| 测序技术 | 实验成本 | 最终成本 | DNA质量要求 | 同源序列的识别能力 | 生物信息学专门知识 | 是否适用于标本材料 |
|---|---|---|---|---|---|---|
| 简化基因组测序 | 高 | 低 | 中-高 | 难 | 中级/高级 | 否 |
| 转录组测序 | 低 | 低 | 中-高 | 相对容易 | 中级 | 否 |
| 杂交捕获测序 | 低 | 中 | 低-中 | 容易 | 中级/高级 | 是 |
| 扩增子测序 | 低 | 中 | 低-中 | 容易 | 初级 | 是 |
| 基因组浅层测序 | 低 | 中 | 低-中 | 容易 | 初级 | 是 |
| [1] | 陈士林, 庞晓慧, 罗焜, 姚辉, 韩建萍, 宋经元 (2013). 生物资源的DNA条形码技术. 生命科学 25, 451-459. |
| [2] | 陈永富, 刘华, 陈巧 (2014). 植物鉴别方法研究现状与展望. 世界林业研究 27(4), 18-23. |
| [3] | 崔颖 (2020). 基于深度学习的木兰科属植物识别系统开发. 硕士论文. 北京: 北京林业大学. pp. 414-493. |
| [4] | 高连明 (2015). DNA条形码在生物多样性编目与评价中的应用. 生物多样性 23, 286-287. |
| [5] | 古尔恰兰·辛格 (2016). 植物系统分类学——综合理论及方法. 生物技术通讯 27, 710. |
| [6] |
郭艳萍, 张浩, 赵新钢, 罗海玲, 张英俊 (2021). DNA宏条形码技术在食草动物食性研究中的应用. 生物技术通报 37, 252-260.
DOI |
| [7] | 郭义红, 孙威江, 林伟东, 陈志丹 (2016). 植物DNA条形码鉴定研究进展. 江苏农业科学 44(7), 19-21. |
| [8] | 侯鑫, 刘俊娥, 赵一之, 赵利清 (2006). 基于ITS序列和trnL-F序列探讨小叶锦鸡儿、中间锦鸡儿和柠条锦鸡儿的种间关系. 植物分类学报 44, 126-134. |
| [9] |
李德铢, 曾春霞 (2015). 植物DNA条形码研究展望. 生物多样性 23, 297-298.
DOI |
| [10] | 李海文, 刘冰, 赖阳均, 杨永, 李奕 (2016). 交互式检索表提升分类学研究与应用效率: 以兰科植物为例. 植物科学学报 34, 151-163. |
| [11] | 刘宇婧, 刘越, 黄耀江, 龙春林 (2011). 植物DNA条形码技术的发展及应用. 植物资源与环境学报 20, 74-82, 93. |
| [12] | 马金双 (2014). 中国植物分类学的现状与挑战. 科学通报 59, 510-521. |
| [13] | 马英, 鲁亮 (2010). DNA条形码技术研究新进展. 中国媒介生物学及控制杂志 21, 275-280. |
| [14] | 毛亚文, 陈江华 (2018). DNA测序技术的发展进程. 亚热带植物科学 47, 94-100. |
| [15] |
宁淑萍, 颜海飞, 郝刚, 葛学军 (2008). 植物DNA条形码研究进展. 生物多样性 16, 417-425.
DOI |
| [16] | 唐建阳, 周先治 (2009). 植物DNA条形码研究现状及应用前景. 中国农学通报 25(24), 35-43. |
| [17] | 王瑞江 (2022). 中国植物分类及标本采集史简述. 广西植物 42, 1-10. |
| [18] | 肖金花, 肖晖, 黄大卫 (2004). 生物分类学的新动向——DNA条形编码. 动物学报 50, 852-855. |
| [19] | 徐疏梅 (2018). 新一代DNA测序技术的应用与研究进展. 徐州工程学院学报(自然科学版) 33(4),60-64. |
| [20] |
闫化学, 于杰 (2010). DNA条形码技术在植物中的研究现状. 植物学报 45, 102-108.
DOI |
| [21] | 杨永, 杨智, 段一凡, 方炎明 (2022). 标本组学——树木学研究的新方法. 南京林业大学学报(自然科学版) 46, 1-6. |
| [22] |
Alsos IG, Lavergne S, Merkel MKF, Boleda M, Lammers Y, Alberti A, Pouchon C, Denoeud F, Pitelkova I, Pușcaș M, Roquet C, Hurdu BI, Thuiller W, Zimmermann NE, Hollingsworth PM, Coissac E (2020). The treasure vault can be opened: large-scale genome skimming works well using herbarium and silica gel dried material. Plants 9, 432.
DOI URL |
| [23] |
Bakker FT (2017). Herbarium genomics: skimming and plastomics from archival specimens. Webbia 72, 35-45.
DOI URL |
| [24] | Bakker FT (2018). Herbarium genomics:plant archival DNA explored. In: Lindqvist C, Rajora OP, eds. Paleogenomics: Genome-Scale Analysis of Ancient DNA. Paleogenomics:Genome-Scale Analysis of Ancient DNA. Cham: Springer. pp. 205-224. |
| [25] |
Beck JB, Semple JC (2015). Next-generation sampling: pairing genomics with herbarium specimens provides species-level signal in Solidago (Asteraceae). Appl Plant Sci 3, 1500014.
DOI URL |
| [26] |
Behjati S, Tarpey PS (2013). What is next generation sequencing? Arch Dis Child-Educ Pract Ed 98, 236-238.
DOI URL |
| [27] |
Besnard G, Christin PA, Malé PJG, Lhuillier E, Lauzeral C, Coissac E, Vorontsova MS (2014). From museums to genomics: old herbarium specimens shed light on a C3 to C4 transition. J Exp Bot 65, 6711-6721.
DOI URL |
| [28] | Bieker VC, Martin MD (2018). Implications and future prospects for evolutionary analyses of DNA in historical herbarium collections. Bot Lett 165, 409-418. |
| [29] |
Brach AR, Song H (2005). ActKey: a web-based interactive identification key program. Taxon 54, 1041-1046.
DOI URL |
| [30] |
Brewer GE, Clarkson JJ, Maurin O, Zuntini AR, Barber V, Bellot S, Biggs N, Cowan RS, Davies NMJ, Dodsworth S, Edwards SL, Eiserhardt WL, Epitawalage N, Frisby S, Grall A, Kersey PJ, Pokorny L, Leitch IJ, Forest F, Baker WJ (2019). Factors affecting targeted sequencing of 353 nuclear genes from herbarium specimens spanning the diversity of angiosperms. Front Plant Sci 10, 1102.
DOI PMID |
| [31] | Calabria I, Pedrola L, Berlanga P, Aparisi MJ, Sánchez- Izquierdo D, Cañete A, Cervera J, Millán JM, Castel V (2016). The new challenge in oncology: next-generation sequencing and its application in precision medicine. An Pediatr (Engl Ed) 85, 273. |
| [32] |
CBOL Plant Working Group, Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim KJ, Kress WJ, Schneider H, van Alphen Stahl J, Barrett SCH, van den Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacón J, Clark A, Clarkson JJ, Conrad F, Devey DS, Ford CS, Hedderson TAJ, Hollingsworth ML, Husband BC, Kelly LJ, Kesanakurti PR, Kim JS, Kim YD, Lahaye R, Lee HL, Long DG, Madriñán S, Maurin O, Meusnier I, Newmaster SG, Park CW, Percy DM, Petersen G, Richardson JE, Salazar GA, Savolainen V, Seberg O, Wilkinson MJ, Yi DK, Little DP (2009). A DNA barcode for land plants. Proc Natl Acad Sci USA 106, 12794-12797.
DOI PMID |
| [33] |
Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madriñán S, Petersen G, Seberg O, Jørgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough TG, Kelly L, Wilkinson M (2007). A proposal for a standardised protocol to barcode all land plants. Taxon 56, 295-299.
DOI URL |
| [34] |
Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, Savolainen V (2005). Land plants and DNA barcodes: short-term and long-term goals. Philos Trans Roy Soc London B Biol Sci 360, 1889-1895.
DOI URL |
| [35] | Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5, e8613. |
| [36] |
China Plant BOL Group, Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB, Fu CX, Zeng CX, Yan HF, Zhu YJ, Sun YS, Chen SY, Zhao L, Wang K, Yang T, Duan GW (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA 108, 19641-19646.
DOI URL |
| [37] |
Chomicki G, Renner SS (2015). Watermelon origin solved with molecular phylogenetics including Linnaean material: another example of museomics. New Phytol 205, 526-532.
DOI PMID |
| [38] |
Costion CM, Lowe AJ, Rossetto M, Kooyman RM, Breed MF, Ford A, Crayn DM (2016). Building a plant DNA barcode reference library for a diverse tropical flora: an example from Queensland, Australia. Diversity 8, 5.
DOI URL |
| [39] |
Cowan RS, Chase MW, Kress WJ, Savolainen V (2006). 300,000 species to identify: problems, progress, and prospects in DNA barcoding of land plants. Taxon 55, 611-616.
DOI URL |
| [40] |
Dodsworth S (2015). Genome skimming for next-generation biodiversity analysis. Trend Plant Sci 20, 525-527.
DOI URL |
| [41] |
Dodsworth S, Guignard MS, Christenhusz MJM, Cowan RS, Knapp S, Maurin O, Struebig M, Leitch AR, Chase MW, Forest F (2018). Potential of herbariomics for studying repetitive DNA in angiosperms. Front Ecol Evol 6, 174.
DOI URL |
| [42] |
Ebach MC, Holdrege C (2005a). More taxonomy, not DNA barcoding. BioScience 55, 823-824.
DOI URL |
| [43] | Ebach MC, Holdrege C (2005b). DNA barcoding is no substitute for taxonomy. Nature 434, 697. |
| [44] |
Enan MR, Palakkott AR, Ksiksi TS (2017). DNA barcoding of selected UAE medicinal plant species: a comparative assessment of herbarium and fresh samples. Physiol Mol Biol Plants 23, 221-227.
DOI URL |
| [45] | Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, Percy DM, Hajibabaei M, Barrett SCH (2008). Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One 3, e2802. |
| [46] | Fu CN, Mo ZQ, Yang JB, Cai J, Ye LJ, Zou JY, Qin HT, Zheng W, Hollingsworth PM, Li DZ, Gao LM (2022). Testing genome skimming for species discrimination in the large and taxonomically difficult genus Rhododendron . Mol Ecol Res 22, 404-414. |
| [47] |
Gao T, Yao H, Song JY, Liu C, Zhu YJ, Ma XY, Pang XH, Xu HX, Chen SL (2010). Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethnopharmacol 130, 116-121.
DOI PMID |
| [48] |
Gardner EM, Johnson MG, Pereira JT, Puad ASA, Arifiani D, Sahromi, Wickett NJ, Zerega NJC (2021). Paralogs and off-target sequences improve phylogenetic resolution in a densely sampled study of the breadfruit genus (Artocarpus, Moraceae). Syst Biol 70, 558-575.
DOI URL |
| [49] | Gauthier J, Mouden C, Suchan T, Alvarez N, Arrigo N, Riou C, Lemaitre C, Peterlongo P (2020). DiscoSnp- RAD: de novo detection of small variants for RAD-Seq population genomics. PeerJ 8, e9291. |
| [50] |
Gernandt DS, Dugua XA, Vázquez-Lobo A, Willyard A, Letelier AM, de la Rosa JAP, Piñero D, Liston A (2018). Multi-locus phylogenetics, lineage sorting, and reticulation in Pinus subsection Australes. Am J Bot 105, 711-725.
DOI PMID |
| [51] |
Gostel MR, Kress WJ (2022). The expanding role of DNA barcodes: indispensable tools for ecology, evolution, and conservation. Diversity 14, 213.
DOI URL |
| [52] |
Gutaker RM, Burbano HA (2017). Reinforcing plant evolutionary genomics using ancient DNA. Curr Opin Plant Biol 36, 38-45.
DOI PMID |
| [53] |
Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA (2007). DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet 23, 167-172.
DOI PMID |
| [54] |
Hart ML, Forrest LL, Nicholls JA, Kidner CA (2016). Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon 65, 1081-1092.
DOI URL |
| [55] |
Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003a). Biological identifications through DNA barcodes. Proc Roy Soc B Biol Sci 270, 313-321.
DOI URL |
| [56] | Hebert PDN, Ratnasingham S, DeWaard JR (2003b). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Roy Soc B Biol Sci 270, S96-S99. |
| [57] | Hollingsworth PM, Graham SW, Little DP (2011). Choosing and using a plant DNA barcode. PLoS One 6, e19254. |
| [58] |
Hollingsworth PM, Li DZ, van der Bank M, Twyford AD (2016). Telling plant species apart with DNA: from barcodes to genomes. Philos Trans Roy Soc B Biol Sci 371, 20150338.
DOI URL |
| [59] |
Ji YH, Liu CK, Yang J, Jin L, Yang ZY, Yang JB (2020). Ultra-barcoding discovers a cryptic species in Paris yunnanensis (Melanthiaceae), a medicinally important plant. Front Plant Sci 11, 411.
DOI URL |
| [60] |
Ji YH, Yang J, Landis JB, Wang SY, Jin L, Xie PX, Liu HY, Yang JB, Yi TS (2022). Genome skimming contributes to clarifying species limits in Paris section Axiparis (Melanthiaceae). Front Plant Sci 13, 832034.
DOI URL |
| [61] |
Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJC, Wickett NJ (2016). HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl Plant Sci 4, 1600016.
DOI URL |
| [62] |
Kane N, Sveinsson S, Dempewolf H, Yang JY, Zhang DP, Engels JMM, Cronk Q (2012). Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am J Bot 99, 320-329.
DOI URL |
| [63] | Kress WJ, Erickson DL (2007). A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One 2, e508. |
| [64] |
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005). Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102, 8369-8374.
DOI PMID |
| [65] |
Li DZ, Liu JQ, Chen ZD, Wang H, Ge XJ, Zhou SL, Gao LM, Fu CX, Chen SL (2011). Plant DNA barcoding in China. J Syst Evol 49, 165-168.
DOI URL |
| [66] |
Li XK, Wang B, Han RC, Zheng YC, Yin HBY, Xu L, Zhang JK, Xu BL (2013). Identification of medicinal plant Schisandra chinensis using a potential DNA barcode ITS2. Acta Soc Bot Pol 82, 283-288.
DOI URL |
| [67] |
Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL (2015). Plant DNA barcoding: from gene to genome. Biol Rev 90, 157-166.
DOI URL |
| [68] | Liu ZF, Ci XQ, Li L, Li HW, Conran JG, Li J (2017). DNA barcoding evaluation and implications for phylogenetic relationships in Lauraceae from China. PLoS One 12, e0175788. |
| [69] |
Meek MH, Larson WA (2019). The future is now: amplicon sequencing and sequence capture usher in the conservation genomics era. Mol Ecol Res 19, 795-803.
DOI URL |
| [70] |
Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA (2007). Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 17, 240-248.
DOI PMID |
| [71] |
Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD (2007). Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol 7, 135.
PMID |
| [72] |
Nazar N, Howard C, Slater A, Sgamma T (2022). Challenges in medicinal and aromatic plants DNA barcoding—lessons from the Lamiaceae. Plants 11, 137.
DOI URL |
| [73] |
Nevill PG, Zhong X, Tonti-Filippini J, Byrne M, Hislop M, Thiele K, van Leeuwen S, Boykin LM, Small I (2020). Large scale genome skimming from herbarium material for accurate plant identification and phylogenomics. Plant Methods 16, 1.
DOI PMID |
| [74] | Osmundson TW, Robert VA, Schoch CL, Baker LJ, Smith A, Robich G, Mizzan L, Garbelotto MM (2013). Filling gaps in biodiversity knowledge for macrofungi: contributions and assessment of an herbarium collection DNA barcode sequencing project. PLoS One 8, e62419. |
| [75] |
Parks M, Cronn R, Liston A (2009). Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol 7, 84.
DOI PMID |
| [76] |
Pennisi E (2007). Wanted: a barcode for plants. Science 318, 190-191.
DOI URL |
| [77] |
Pillon Y, Johansen J, Sakishima T, Chamala S, Barbazuk WB, Roalson EH, Price DK, Stacy EA (2013). Potential use of low-copy nuclear genes in DNA barcoding: a comparison with plastid genes in two Hawaiian plant radiations. BMC Evol Biol 13, 35.
DOI PMID |
| [78] | Rasmussen K, Thyrring J, Muscarella R, Borchsenius F (2017). Climate-change-induced range shifts of three allergenic ragweeds (Ambrosia L.) in Europe and their potential impact on human health. PeerJ 5, e3104. |
| [79] |
Reichelt N, Wen J, Pätzold C, Appelhans MS (2021). Target enrichment improves phylogenetic resolution in the genus Zanthoxylum (Rutaceae) and indicates both incomplete lineage sorting and hybridization events. Ann Bot 128, 497-510.
DOI URL |
| [80] |
Ren C, Wang L, Nie ZL, Johnson G, Yang QE, Wen J (2021). Development and phylogenetic utilities of a new set of single-/low-copy nuclear genes in Senecioneae (Asteraceae), with new insights into the tribal position and the relationships within subtribe Tussilagininae. Mol Phylogene Evol 162, 107202.
DOI URL |
| [81] |
Ruhsam M, Rai HS, Mathews S, Ross TG, Graham SW, Raubeson LA, Mei WB, Thomas PI, Gardner MF, Ennos RA, Hollingsworth PM (2015). Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol Ecol Res 15, 1067-1078.
DOI URL |
| [82] | Ruppert KM, Kline RJ, Rahman MS (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Glob Ecol Conserv 17, e00547. |
| [83] |
Sablok G, Amiryousefi A, He XL, Hyvönen J, Poczai P (2019). Sequencing the plastid genome of giant ragweed (Ambrosia trifida, Asteraceae) from a herbarium specimen. Front Plant Sci 10, 218.
DOI URL |
| [84] |
Sánchez CD, Richardson JE, Hart M, Serrano J, Cárdenas D, Gonzalez MA, Cortés BR (2022). A plea to DNA barcode type specimens: an example from Micropholis (Sapotaceae). Taxon 71, 154-167.
DOI URL |
| [85] |
Sarmashghi S, Bohmann K, Gilbert MTP, Bafna V, Mirarab S (2019). Skmer: assembly-free and alignment-free sample identification using genome skims. Genome Biol 20, 34.
DOI PMID |
| [86] |
Sebastian P, Schaefer H, Telford IRH, Renner SS (2010). Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci USA 107, 14269-14273.
DOI PMID |
| [87] | Staats M, Erkens RHJ, van de Vossenberg B, Wieringa JJ, Kraaijeveld K, Stielow B, Geml J, Richardson JE, Bakker FT (2013). Genomic treasure troves: complete genome sequencing of herbarium and insect museum specimens. PLoS One 8, e69189. |
| [88] |
Straub SCK, Fishbein M, Livshultz T, Foster Z, Parks M, Weitemier K, Cronn RC, Liston A (2011). Building a model: developing genomic resources for common milkweed (Asclepias syriaca) with low coverage genome sequencing. BMC Genomics 12, 211.
DOI PMID |
| [89] |
van de Paer C, Hong-Wa C, Jeziorski C, Besnard G (2016). Mitogenomics of Hesperelaea, an extinct genus of Oleaceae. Gene 594, 197-202.
DOI URL |
| [90] | Vijayan K, Tsou CH (2010). DNA barcoding in plants: taxonomy in a new perspective. Curr Sci 99, 1530-1541. |
| [91] |
Villaverde T, Pokorny L, Olsson S, Rincón-Barrado M, Johnson MG, Gardner EM, Wickett NJ, Molero J, Riina R, Sanmartín I (2018). Bridging the micro- and macro-a)evolutionary levels in phylogenomics: Hyb-seq solves relationships from populations to species and above. New Phytol 220, 636-650.
DOI PMID |
| [92] | Wang J, Fu CN, Mo ZQ, Möller M, Yang JB, Zhang ZR, Li DZ, Gao LM (2022). Testing the complete plastome for species discrimination, cryptic species discovery and phylogenetic resolution in Cephalotaxus ( Cephalotaxaceae). Front Plant Sci 13, 768810. |
| [93] |
Weitemier K, Straub SCK, Cronn RC, Fishbein M, Schmickl R, McDonnell A, Liston A (2014). Hyb-seq: combining target enrichment and genome skimming for plant phylogenomics. Appl Plant Sci 2, 1400042.
DOI URL |
| [94] |
Welch AJ, Collins K, Ratan A, Drautz-Moses DI, Schuster SC, Lindqvist C (2016). The quest to resolve recent radiations: plastid phylogenomics of extinct and endangered Hawaiian endemic mints (Lamiaceae). Mol Phylogene Evol 99, 16-33.
DOI URL |
| [95] |
Xu T, Kong LF, Li Q (2022). Testing efficacy of assembly-free and alignment-free methods for species identification using genome skims, with Patellogastropoda as a test case. Genes 13, 1192.
DOI URL |
| [96] | Yu XQ, Jiang YZ, Folk RA, Zhao JL, Fu CN, Fang L, Peng H, Yang JB, Yang SX (2022). Species discrimination in Schima (Theaceae): next-generation super-barcodes meet evolutionary complexity. Mol Ecol Res 22, 3161-3175. |
| [97] |
Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, Yang JB (2018). Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods 14, 43.
DOI |
| [98] |
Zhang L, Huang YW, Huang JL, Ya JD, Zhe MQ, Zeng CX, Zhang ZR, Zhang SB, Li DZ, Li HT, Yang JB (2023). DNA barcoding of Cymbidium by genome skimming: call for next-generation nuclear barcodes. Mol Ecol Res 23, 424-439.
DOI URL |
| [99] |
Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS (2017). Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol 214, 1355-1367.
DOI URL |
| [100] |
Zhang W, Sun YZ, Liu J, Xu C, Zou XH, Chen X, Liu YL, Wu P, Yang XY, Zhou SL (2021). DNA barcoding of Oryza: conventional, specific, and super barcodes. Plant Mol Biol 105, 215-228.
DOI |
| [101] |
Zhou XG, Ren LF, Li YT, Zhang M, Yu YD, Yu J (2010). The next-generation sequencing technology: a technology review and future perspective. Sci China Life Sci 53, 44-57.
DOI PMID |
| [1] | 惠城阳, 章巧依, 刘腾腾, 刘维勇, 周丽娜, 金鑫杰, 张永华, 刘金亮. 温州大罗山主要植被类型及物种组成特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [2] | 杜燕 刘鑫 张瀚曰 马少伟 包维楷. 中国高山松群系的群落特征[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [3] | 韩鹏宾 何斌 陆树刚 唐勤. 中国柔毛油杉林的群落类型及特征[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [4] | 何斌 韩鹏宾 肖书礼 裴康迪 唐勤. 铁坚油杉林的群落类型及特征[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [5] | 韩鹏宾 裴康迪 何斌 肖书礼 唐勤. 中国云南铁杉林的群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [6] | 张坤, 伊丽晴, 刘占军, 张志杰, 王顼, 李明乐, 苗百岭, 张景慧, 李智勇, 董雷, 梁存柱. 毛刺锦鸡儿群落特征及分类[J]. 植物生态学报, 2025, 49(植被): 0-. |
| [7] | 郭欢敏, 沈小雪, 李瑞利. 深圳湾福田红树林自然保护区物种共存特征与物种分布概率研究[J]. 植物生态学报, 2025, 49(预发表): 1-. |
| [8] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [9] | 白帆, 王杨. 2007–2015年北京东灵山暖温带落叶阔叶林长期监测样地植物物种组成和群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 1-. |
| [10] | 蔡榕榕, 沈历都, 刘雅各, 费雯丽, 戴冠华. 2005-2010年长白山阔叶红松林长期监测样地的物种组成和群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 1-. |
| [11] | 刘世忠 张倩媚 张德强 刘菊秀 褚国伟 李跃林. 1999-2015年鼎湖山季风常绿阔叶林长期监测样地的植物物种组成和群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 1-0. |
| [12] | 饶兴权 蔡锡安 林永标 刘素萍. 2005-2015年鹤山站马占相思林长期监测样地的物种组成和群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [13] | 郑立媛, 徐茜竹, 尹嘉淇, 孙小雯, 王艳. 沈阳城郊近河农田退耕地野大豆群落生态位和种间联结研究[J]. 植物生态学报, 2025, 49(濒危植物的保护与恢复): 1-. |
| [14] | 李利平, 余堃, 陈芳淼, 陈强, 景雨航, 牛振国, 胡辰璐, 覃海宁, 黄慧萍, 田亦陈. 基于IUCN全球评估数据的中国高等植物红色名录指数[J]. 植物生态学报, 2025, 49(濒危植物的保护与恢复): 1-. |
| [15] | 韦丹丹, 杜燕, 包维楷, 胡斌, 张瀚曰, 王瀚婕, 唐圆圆, 黄龙, 郭昌安, 刘鑫. 冬麻豆群落的地理分布、特征和分类[J]. 植物生态学报, 2025, 49(濒危植物的保护与恢复): 1-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||