蒙古冰草(即沙芦草Agropyron mongolicum))是禾本科多年生二倍体草本植物, 茎叶柔软, 根系高度发达, 返青早, 分蘖能力强, 适应性强, 是干旱草原上优良的多年生牧草, 在我国天然牧场和人工草地播种中发挥重要作用。目前, 蒙古冰草等优质牧草广泛种植于我国中西部荒漠草原, 也作为生态修复草用于寒冷和干旱地区的生态恢复, 是牧草生产和生态建设中的重要植物资源(林克剑等, 2023)。此外, 蒙古冰草作为小麦(Triticum aestivum)的野生近缘种, 为小麦等禾本科作物育种提供了重要的抗性基因资源, 对研究物种起源与演化及遗传多样性具有重要意义(Che and Li, 2007)。
木质素是一种复杂的多酚聚合物, 由单木酚通过苯丙烷途径合成。木质素聚合物主要包括对羟苯基木质素、愈创木基木质素和紫丁香基木质素3种类型(邱锐等, 2023)。目前已知有12种酶参与木质素合成, 其中肉桂醇脱氢酶(cinnamyl alcohol dehydrogenase, CAD)可在NADPH作为辅助因子存在时还原松柏醛、芥子醛和对香豆醛, 产生松柏醇、芥子醇和对香豆醇等木质素单体, 是参与木质素单体合成的关键酶(李桢等, 2009)。CAD广泛存在于高等植物基因组中, 参与干旱和盐胁迫等过程, 与植物抗逆性密切相关(Ibrahim et al., 2019)。Janiak等(2016)研究表明, LuCAD6参与植物的干旱胁迫反应, 导致亚麻(Linum usitatissimum)在长期干旱条件下根部积累木质素。在盐胁迫下, 棉花(Gossypium hirsutum) CAD活性发生改变, 在干旱和盐组合胁迫下, CAD转录水平显著升高(Ibrahim et al., 2019)。全基因组分析表明, 植物中CAD基因以家族形式存在, 研究人员分别在甜瓜(Cucumis melo)、苜蓿(Medicago sativa)、水稻(Oryza sativa)和高粱(Sorghum bicolor)等物种中鉴定到5、7、12和14个不同的CAD序列(Tobias and Chow, 2005; Zhang et al., 2006; Saballos et al., 2009; Jin et al., 2014)。系统发育分析表明, 这些CAD基因可分为3-7个亚家族(Ma, 2010; Bukh et al., 2012)。亚家族I中的CAD成员被认为与植物木质素生物合成相关。在拟南芥(Arabidopsis thaliana)中, AtCAD4和AtCAD5是参与木质素单体生物合成过程的主要CAD (Sibout et al., 2003)。在水稻CAD家族中, OsCAD2是唯一属于亚家族I的CAD, 参与木质素生物合成(Barakat et al., 2009)。然而, 蒙古冰草的CAD基因家族及其功能鉴定至今尚未见报道。
随着全球气温升高, 干旱正在成为植物生长发育的主要威胁之一(Rollins et al., 2013)。干旱胁迫通过扰乱水分平衡和降低水分利用效率在植物细胞中引起复杂的生理生化反应。蒙古冰草具有耐旱、耐低温、耐盐碱、抗小麦锈病及其它病原菌等优良特性, 因此挖掘蒙古冰草抗逆基因对于牧草分子育种具有重要意义。赵彦等(2015)在蒙古冰草中鉴定到MwDREB3基因, 发现其对低温、干旱、高盐和ABA诱导具有一定的响应。轻度盐胁迫对蒙古冰草幼苗生长有一定的促进作用, 可通过提高酶活性维持活性氧产生与清除之间的平衡, 使膜通透性降低。而中度和重度盐胁迫对幼苗生长具有明显的抑制作用(王荣华等, 2003, 2004)。对蒙古冰草NAC家族基因的深入分析也表明, AmNAC100和AmNAC102-2可能参与干旱胁迫响应, 具有调控植物生长发育及响应非生物逆境等多种功能(范菠菠等, 2021)。近年来, 多种植物中的研究表明, CAD基因对植物抗逆性具有重要作用。干旱胁迫诱导甜瓜3个CAD基因(即CmCAD1、CmCAD2和CmCAD3)的表达, 以促进木质素的生物合成(Xu et al., 2011)。对毛竹(Phyllostachys edulis) PheCAD的研究发现, 经非生物胁迫处理后, PheCAD2被显著诱导表达(Vasupalli et al., 2021)。上述结果表明, CAD基因既参与木质化, 也响应非生物胁迫(Liu et al., 2020)。因此, 深入挖掘蒙古冰草CAD基因可为禾本科作物育种提供新的且有应用前景的基因资源。
本研究通过对蒙古冰草全长转录组序列进行深入挖掘, 筛选到12个CAD候选基因。系统进化分析和结构预测表明, AmCAD基因可分为4个亚家族, 具有典型的2个Zn2+和NADPH结构域, 并鉴定到AmCAD为参与木质素合成的候选基因。该基因在茎秆等木质化程度较高的组织部位高表达; 体外酶活分析也表明, AmCAD重组蛋白对松柏醛、芥子醛及对香豆醛均具有很强的催化能力。此外, qRT-PCR分析表明, AmCAD的表达在干旱胁迫下受到显著诱导。综上所述, 本研究在对蒙古冰草CAD基因家族进行生物信息学分析的基础上, 鉴定到参与木质素合成并响应干旱胁迫的关键基因AmCAD。以该基因为靶点在蒙古冰草中进行深入研究, 对于提高牧草品质及增强牧草逆境胁迫耐受性具有重要意义。
1 材料与方法
1.1 蒙古冰草CAD基因的筛选和分析
从拟南芥(Arabidopsis thaliana L.)信息资源中心(
1.2 蒙古冰草CAD蛋白的理化性质及保守基序分析
利用TBtools分析AmCAD氨基酸分子量、等电点、不稳定系数、亲水指数以及脂溶指数。利用WoLF PSORT (
1.3 蒙古冰草CAD基因家族的系统发育分析
利用MEGA7 (
1.4 蒙古冰草CAD蛋白三维结构模型预测
使用SWISS-MODEL (
1.5 AmCAD序列扩增和载体构建
根据AmCAD的cDNA序列设计引物AmCAD-pET- 32a-F: 5'-ATGGGCAGCGTCGACGCCTCC-3'; AmCAD-pET32a-R: 5'-TCAGGCGGCGTTCTCGATGTTG-3'。以蒙古冰草cDNA为模板扩增获得PCR产物。用EcoRI (NEB)酶切pET-32a质粒, 将PCR扩增片段的胶回收产物在infusion连接酶(诺唯赞生物科技股份有限公司, 南京)作用下连接到酶切后的pET-32a载体上, 冰浴后热激转入DH5α感受态细胞, 通过菌液PCR筛选阳性克隆(检测引物序列为F: 5'-CGAACGCCAGCACATGGACAGC-3'; R: 5'-GACCCGTTTAGAGGCCCCAAGG-3')并对阳性克隆进行测序验证, 测序引物同PCR检测引物。
1.6 AmCAD重组蛋白的高效表达和纯化
用热激法将包含pET-32a_AmCAD的质粒转入BL21感受态细胞, 通过菌液PCR筛选阳性克隆。将阳性菌株接种到添加羧苄的LB液体培养基中, 37°C预培养过夜, 按1:100的比例转接到50 mL LB液体培养基中, 培养至OD600=0.6时开始诱导蛋白表达, 经IPTG过夜诱导后收集菌体, 超声破碎后, 采用层析法在Ni2+柱中纯化上清液中的AmCAD重组蛋白, 收集上清液粗蛋白、滤出液和纯化蛋白等。参照Ma和Tian (2005)的方法进行SDS-PAGE检测。
1.7 AmCAD重组蛋白的酶学性质
以对香豆醛、松柏醛和芥子醛为底物, 用高效液相色谱法(HPLC)检测AmCAD重组蛋白的催化活性。酶活反应体系为: 20 μL Tris-HCL, 238 μL H2O, 10 μL NADPH (2 mmol∙L-1), 20 μL纯化的重组AmCAD蛋白, 12 μL底物(2 mmol∙L-1)。30°C水浴反应30分钟, 加入甲醇溶液终止反应, 过滤后进行液相检测。同时, 以高温灭活的重组蛋白替代活性蛋白, 为每种底物增加1个空白对照。
为比较AmCAD重组蛋白对不同底物的最适催化条件, 在反应温度为25°C、30°C、37°C、45°C和55°C下进行酶活检测以确定最适反应温度, 继而在最适反应温度下调节缓冲液pH至3、4.5、6.5、7.5和9, 分别进行酶活检测以确定最适反应pH。
通过双倒数作图法(Lineweaver-Burk plot)对AmCAD重组蛋白的动力学常数进行测定。以初始速率(V0)和底物浓度(S)为坐标, 根据米氏方程V0=Vmax S/(Km+S)进行非线性拟合得出米氏常数(Km)和最大反应速率(Vmax), 并计算AmCAD重组蛋白的催化常数Kcat。以上所有实验均设3次重复。
1.8 AmCAD基因在不同组织中的表达量
样品取自内蒙古农业大学2018年种植的蒙古冰草蒙农一号, 分别取根、茎、叶、叶鞘和幼穗磨样, 用TransZol法(全式金生物公司, Cat No.ET101-01)提取嫩叶总RNA, 然后反转录为cDNA (全式金生物公司试剂盒, Cat No.AE311)。通过qRT-PCR法检测AmCAD的表达量(引物序列为AmCAD_qF: 5'-CTCG- TGCTGATGGGCGTGAT-3'; AmCAD_qR: 5'-CCGA- TGAAGCTGCCCGTGAT-3')。参照黄文华(2014)的报道, 以蒙古冰草18SrRNA作为内参基因(18SrRNA-F: 5'-CAATGGGAAGCAAGGCTGTAA-3'; 18SrRNA-R: 5'-AACAATCCGAACTGAGGCAATC-3')。反应程序为: 95°C30秒, 95°C5秒, 60°C30秒, 共35个循环。采用2-ΔΔCT法计算各基因的相对表达量(Livak and Schmittgen, 2001)。
1.9 AmCAD基因在干旱胁迫下的表达量
在滤纸上萌发蒙古冰草种子, 2周后取长势一致的蒙古冰草置于1/4Hoglands营养液中预培养3天。然后在溶液中添加甘露醇, 使终浓度分别达75、150和250 mmol∙L-1, 以1/4Hoglands营养液水培作为对照, 模拟干旱处理。分别在处理后第0、2、6和12小时进行根茎叶混合取样, 样品用液氮速冻后研磨, 使用Trizol法提取RNA, 然后反转录为cDNA, 采用qRT- PCR分析AmCAD基因在不同处理时间及不同甘露醇浓度下的表达量。
2 结果与讨论
2.1 AmCAD基因家族成员的鉴定
利用拟南芥CAD基因序列在蒙古冰草全长转录组本地数据库中进行比对, 共筛选到69条序列, 其中12条序列具有完整的保守结构域, 可作为候选AmCAD序列用于进一步分析。AmCAD基因编码蛋白质的氨基酸数目介于269-421之间, 其中, PB.47633.2相对分子质量最大(44.35 kDa), PB.62308.1相对分子质量最小(28.62 kDa)。蒙古冰草CAD蛋白具有相似的等电点(pI=5.40-6.66)、不稳定系数(23.05-31.84)和脂溶指数(83.80-93.88) (表1)。同时, 除PB.82665.1外, AmCAD蛋白亲水指数均大于0, 为疏水性蛋白。亚细胞定位预测结果表明, 不同AmCAD蛋白的定位不同, PB.80567.1定位于过氧化物酶体, PB.47633.2、PB.55683.1和PB.84043.1定位于叶绿体, 其余8个AmCAD蛋白定位于细胞质。
表1 蒙古冰草AmCAD蛋白理化性质分析
Table 1
| Sequence ID | Number of amino acid | Molecular weight (Da) | Theoretical pI | Instability index | Aliphatic index | Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| PB.70108.1 | 360 | 38591.48 | 5.87 | 23.05 | 90.33 | 0.025 |
| PB.82665.1 | 354 | 38437.86 | 6.66 | 24.94 | 84.75 | -0.098 |
| PB.80567.1 | 358 | 38685.52 | 6.60 | 30.79 | 89.27 | 0.012 |
| PB.80906.1 | 355 | 38256.89 | 5.68 | 31.84 | 88.42 | 0.037 |
| PB.62308.1 | 269 | 28625.35 | 6.31 | 31.41 | 92.45 | 0.166 |
| PB.17482.1 | 344 | 36802.76 | 5.86 | 31.22 | 86.13 | 0.103 |
| PB.52032.1 | 373 | 38747.35 | 6.53 | 24.05 | 91.58 | 0.136 |
| PB.47633.2 | 421 | 44353.00 | 6.42 | 31.04 | 84.28 | 0.092 |
| PB.55683.1 | 421 | 44338.97 | 6.42 | 29.60 | 83.80 | 0.088 |
| PB.83678.1 | 356 | 37359.82 | 5.40 | 25.49 | 92.25 | 0.146 |
| PB.84043.1 | 356 | 37036.55 | 5.84 | 26.83 | 93.88 | 0.211 |
| PB.84184.1 | 356 | 37019.73 | 6.08 | 23.61 | 92.25 | 0.195 |
2.2 AmCAD蛋白结构域分析
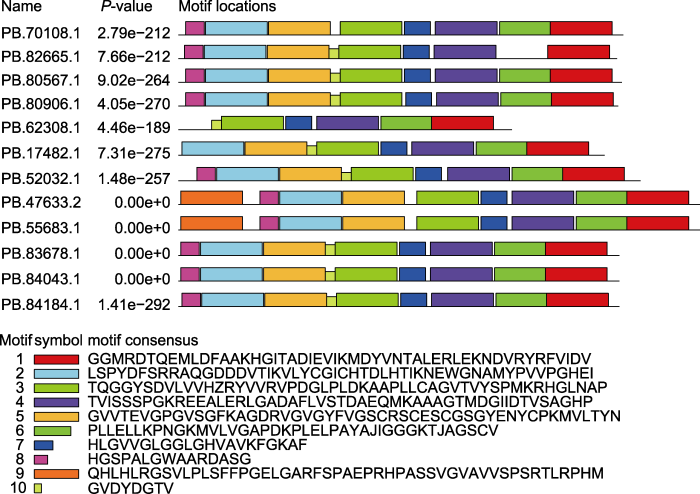
为明确不同AmCAD蛋白基序是否具有相似性, 利用MEME软件预测蛋白结构域, 使用TBtools绘图。结果表明, AmCAD蛋白结构较为保守, 共鉴定到9个保守基序(分别命名为Motif1-9), 分别用不同的颜色表示(图1)。PB.47633.2和PB.55683.1的motif数量最多, 包含全部预测到的9个motif。而PB.62308.1的motif最少, 仅包含Motif1、Motif3、Motif4、Motif6和Motif7这5个保守基序。AmCAD蛋白结构域核心基序的保守性和蛋白结构的多样性预示着蒙古冰草CAD基因功能的多样性。
图1
图1
蒙古冰草AmCAD蛋白的基序分析
Figure 1
Motif analysis of AmCAD protein in Agropyron mongolicum
2.3 AmCAD蛋白家族系统进化分析
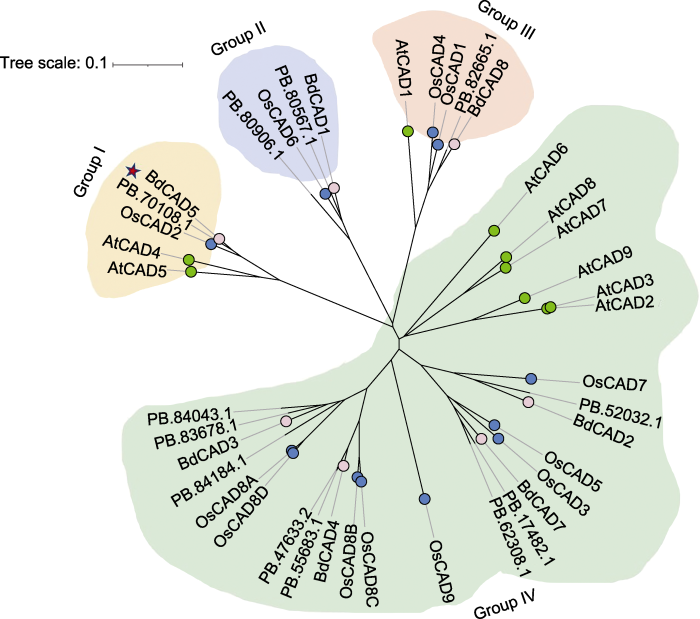
为确定AmCAD与其它高等植物CAD的进化关系, 利用3个物种的28个CAD氨基酸序列(二穗短柄草7个, 拟南芥9个, 水稻12个)与鉴定到的12个蒙古冰草CAD氨基酸序列构建系统进化树。结果表明, 12个AmCAD可划分为4个亚家族, 命名为Group I-IV。其中, AmCAD主要聚类在Group IV中(8个), 而Group I、Group II和Group III中分别包含1、2和1个AmCAD (图2)。
图2
图2
蒙古冰草与拟南芥、水稻和二穗短柄草CAD的系统进化树
红色五角星代表可能参与木质素合成的蒙古冰草CAD蛋白; 粉色点代表二穗短柄草CAD蛋白; 绿色点代表拟南芥CAD蛋白; 蓝色点代表水稻CAD蛋白。
Figure 2
Phylogenetic tree of CADs in Agropyron mongolicum, Arabidopsis thaliana, Oryza sativa, and Brachypodium distachyon
The red pentagram represents the functional CAD protein of A. mongolicum that may be involved in lignin synthesis; the pink dots represent the CAD protein of B. distachyon; the green dots represent the CAD protein of A. thaliana; and the blue dots represent the CAD protein of O. sativa.
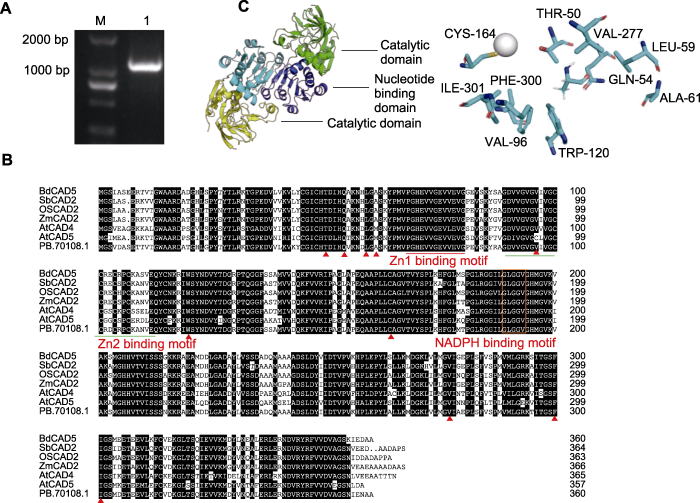
2.4 AmCAD序列扩增及蛋白结构分析
我们首先从蒙古冰草cDNA中成功扩增了AmCAD编码序列(图3A)。为进一步确定AmCAD与木质素单体合成的相关性, 将AmCAD与其它植物中参与木质素单体合成的CAD进行氨基酸序列比对分析(图3B)。结果表明, 所有具催化活性的CAD蛋白均含有2个与Zn2+催化相关的结构域: Zn1结合基序GHE(X)2G(X)5 G(X)2V和Zn2结合基序GD(X)10C(X)2C(X)2C(X)7C (McKie et al., 1993)以及1个与NADPH结合相关的富含甘氨酸的保守结构域GLGGLGGV(L)G (Saballos et al., 2009)。基于拟南芥AtCAD5 (PDB:2cf5)晶体结构, 我们对AmCAD进行三维结构建模分析(图3C)。结果表明, AmCAD为同源二聚体结构, 包括催化结构域和核酸结构域。构成AmCAD底物结合口袋的包括10个氨基酸残基, 分别为TRP120、VAL96、ALA61、GLN54、CYS164、LEU59、THR50、ILE301、PHE300和VAL277。共同的结构基础表明, AmCAD可能是具有肉桂醛类底物催化能力的功能蛋白。
图3
图3
蒙古冰草AmCAD基因扩增和蛋白结构分析
(A) AmCAD电泳图(M: DNA ladder; 1: 扩增序列); (B) 蒙古冰草与其它物种CAD氨基酸序列比对(构成底物结合口袋的残基用三角形(▲)标记); (C) AmCAD蛋白质三维结构模型和底物结合口袋(底部亚基的核苷酸结合和催化结构域分别呈黄色和蓝色, 在上部亚基中的对应结构分别以绿色和深蓝色显示, 锌离子用灰色球体表示)
Figure 3
Amplification of Agropyron mongolicum AmCAD and protein structure analysis
(A)AmCAD gel map (M: DNA ladder; 1: Amplification sequence); (B) Comparison of CAD amino acid sequences among A. mongolicum and other species (the residue forming the substrate binding pocket is marked with a triangle (▲)); (C) Three-dimensional protein structure model and substrate binding pocket of AmCAD (the nucleotide binding and catalytic domains of the bottom subunit are colored yellow and blue, respectively, the corresponding compounds in the upper subunit are colored green and dark blue, while the zinc ions are indicated by gray spheres)
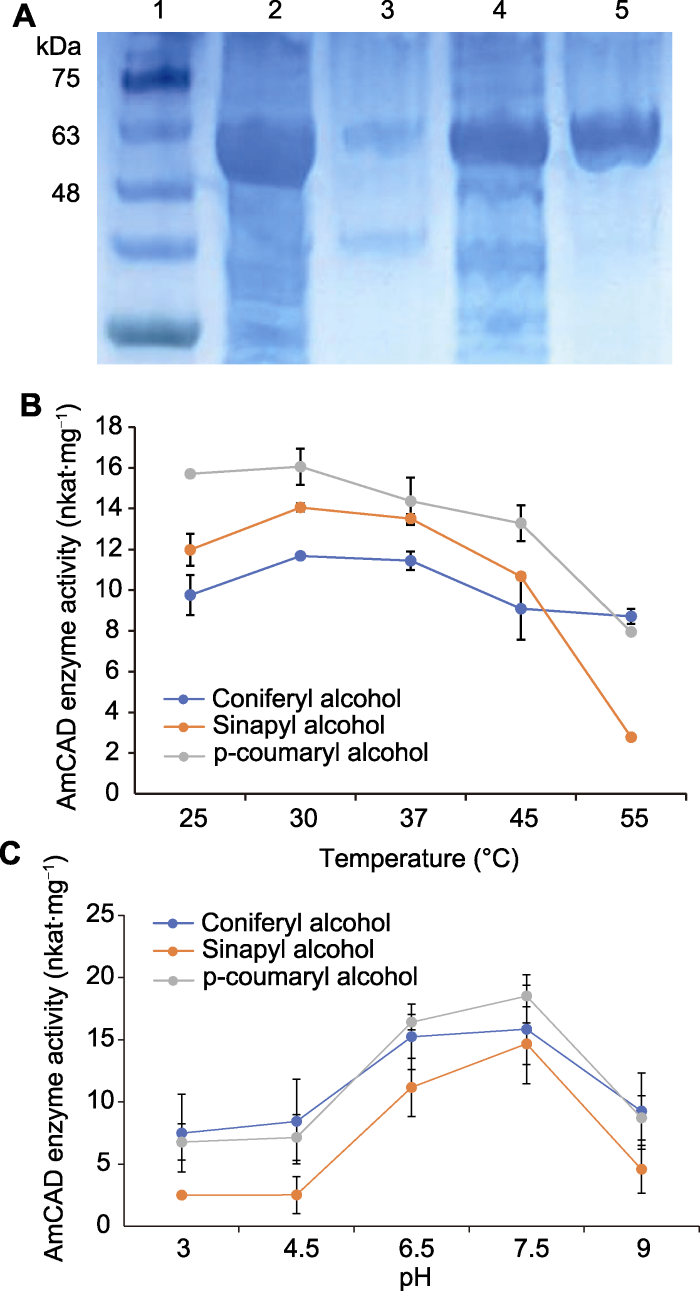
2.5 AmCAD重组蛋白的酶学性质分析
将含有pET-32a_AmCAD质粒的BL21菌株经IPTG过夜诱导, 采用层析法在Ni2+柱中纯化蛋白。聚丙烯酰胺凝胶电泳(SDS-PAGE)结果表明, AmCAD重组蛋白的分子量约为55.6 kDa (图4A)。
图4
图4
AmCAD重组蛋白在大肠杆菌中的异源表达(A)及不同温度(B)和pH (C)下的酶学性质
1: 蛋白Marker; 2: 上清液; 3: 沉淀; 4: 流出液; 5: 纯化蛋白
Figure 4
Heterologous expression (A) and enzymatic properties of AmCAD recombinant protein in Escherichia coli at different temperatures (B) and pH (C)
1: Marker; 2: Supernatant; 3: Precipitant; 4: Effluent; 5: Purified protein
AmCAD重组蛋白在不同温度和pH下的酶活比较分析表明, 该蛋白催化不同底物的最佳反应体系均为pH7.5和温度30°C (图4B, C)。在25-45°C下, AmCAD对对香豆醛的还原活性最高, 在pH3.5-7.5的缓冲液中, AmCAD对对香豆醛和松柏醛的还原活性相当且均大于芥子醛。上述结果表明, AmCAD重组蛋白具有较高的温度稳定性和较强的pH敏感性。
为探明AmCAD重组蛋白对不同底物的偏好性, 进而通过分子设计手段获得具有不同木质素结构的蒙古冰草新种质。本研究在最适反应条件下(30°C, pH7.5)测定了AmCAD重组蛋白对对香豆醛、芥子醛和松柏醛等底物的酶动力学参数(表2)。结果表明, 以松柏醛为底物时, AmCAD的Km为8.94 μmol·L-1, Vmax为6.78 μmol·L-1·s-1, Kcat为441.37·s-1, Kcat/Km为49.35 μmol·L-1·s-1。以芥子醛为底物时, AmCAD的Km为10.35 μmol·L-1, Vmax为7.18 μmol·L-1·s-1, Kcat为461.51·s-1, Kcat/Km为45.26 μmol·L-1·s-1。以对香豆醛为底物时, AmCAD的Km为35.35 μmol·L-1, Vmax为15.21 μmol·L-1·s-1, Kcat为977.60·s-1, Kcat/Km为27.65 μmol·L-1·s-1。可以看出, AmCAD对松柏醛的亲和力最强, 芥子醛次之, 对香豆醛最弱。Kcat/Km值分析结果显示, AmCAD还原松柏醛的效率是还原对香豆醛的2倍。上述结果表明, AmCAD重组蛋白对不同肉桂醛类底物具有较强的选择性。
表2 AmCAD重组蛋白催化不同底物的酶动力学特性
Table 2
| Substract | Km (μmol·L-1) | Vmax (μmol·L-1·s-1) | Kcat (·s−1) | Kcat/Km (μmol·L-1·s-1) |
|---|---|---|---|---|
| Coniferylaldehyde | 8.94±0.15 | 6.87±0.008 | 441.37±0.53 | 49.35±0.90 |
| Sinapaldehyde | 10.35±2.14 | 7.18±0.43 | 461.51±27.94 | 45.26±6.69 |
| p-coumaraldehyde | 35.35±3.74 | 15.21±1.60 | 977.60±102.97 | 27.65±0.01 |
Km: 米氏常数; Vmax: 最大反应速率; Kcat: 催化常数
Km: Michaelis constant; Vmax: Maximum reaction rate; Kcat: Catalytic constant
2.6 AmCAD在不同组织中的表达量分析
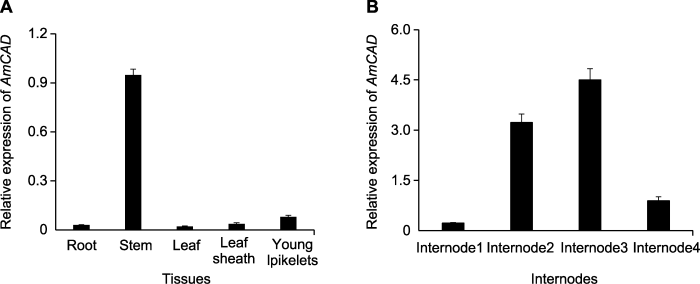
为进一步探明AmCAD基因的组织表达特性, 在蒙古冰草抽穗期分别对根、茎、叶、叶鞘和幼穗取样, 并分析不同组织中AmCAD的表达量。结果表明, AmCAD基因表达存在组织特异性, 在茎中的表达量最高, 其次是幼穗和叶鞘, 在根和叶中的表达量极低(图5A)。
图5
图5
AmCAD在蒙古冰草不同组织(A)和茎节部位(B)的表达量
Figure 5
Expression levels of AmCAD in different tissues (A) and internodes (B) of Agropyron mongolicum
为进一步分析该基因在茎节中的表达模式, 选择抽穗期蒙古冰草的茎节, 按照从幼穗到根的取样顺序分为4个部位, 分别命名为茎节1 (I1)、茎节2 (I2)、茎节3 (I3)和茎节4 (I4)。结果表明, 蒙古冰草AmCAD基因在I3中表达量最高, 在I1中表达量最低, 且在I3中表达量约为I1的4倍(图5B)。AmCAD在蒙古冰草茎中的特异表达以及在不同茎节中表达量的差异性, 表明该基因在蒙古冰草木质素合成和生长发育中可能发挥重要作用。
2.7 AmCAD在模拟干旱胁迫下的表达分析
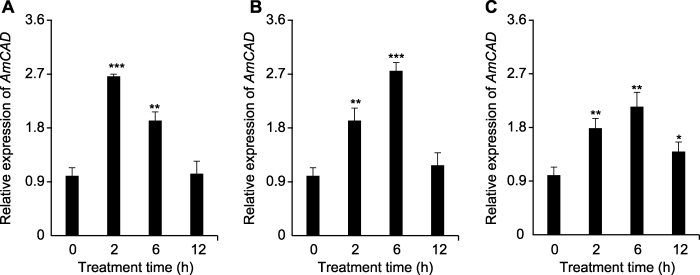
为探明AmCAD基因在干旱胁迫下的表达模式, 用不同浓度甘露醇模拟干旱胁迫, 分别在处理后第0、2、6和12小时取样进行qRT-PCR分析。结果表明, 干旱胁迫显著诱导蒙古冰草AmCAD的表达(图6)。在75、150和250 mmol·L-1甘露醇处理下, AmCAD的表达量最高被诱导约2.5、2.7和2倍。此外, AmCAD表达量在不同浓度甘露醇处理下均呈现先升高后降低的趋势, 并在处理12小时后基本恢复正常表达水平。AmCAD在不同浓度甘露醇处理下都能被快速诱导表达, 说明AmCAD可能在蒙古冰草对干旱胁迫抗性中发挥功能, 可作为作物抗逆性改良的潜在靶基因, 因此具有一定的应用价值。
图6
图6
AmCAD基因在不同处理时间及不同浓度甘露醇模拟干旱胁迫下的表达分析
(A) 75 mmol∙L-1甘露醇处理; (B) 150 mmol∙L-1甘露醇处理; (C) 250 mmol∙L-1甘露醇处理。* P<0.05; ** P<0.01; *** P<0.001
Figure 6
Expression analysis of AmCAD gene under drought stress at different time points and mannitol concentrations
(A) 75 mmol∙L-1 mannitol treatment; (B) 150 mmol∙L-1 mannitol treatment; (C) 250 mmol∙L-1 mannitol treatment. * P<0.05; ** P<0.01; *** P<0.001
2.8 讨论
肉桂醇脱氢酶是参与植物次生代谢产物尤其是木质素生物合成过程的关键酶, 在植物生长发育和对生物/非生物胁迫的耐受性方面发挥重要作用(Lange et al., 1995)。前人系统分析了拟南芥(Sibout et al., 2003)、杨树(Populus) (Barakat et al., 2009)、水稻(Tobias and Chow, 2005)和二穗短柄草(Bukh et al., 2012)等物种中的CAD基因家族, 分别鉴定到9、15、12和7个CAD基因。本研究以蒙古冰草全长转录组为基础, 共筛选到12个CAD序列, 在数量上与上述已鉴定的物种相近。所有鉴定到的AmCAD在蛋白结构和分子特征上表现出一定的保守性, 除PB.623.08.1外, 其它CAD序列均包含至少7个保守基序。预测结果表明, 鉴定到的所有AmCAD等电点均在5.40-6.66之间。亚细胞定位预测结果表明, 蒙古冰草CAD多定位于细胞质, 仅在PB.47633.2、PB.55683.1、PB.84043.1以及PB.80567.1中发现叶绿体和过氧化物酶体定位信号肽, 这在前人的研究中也有报道(Bukh et al., 2012; Jin et al., 2014; Shafiei et al., 2023)。在植物中, CAD具有调控生长发育和影响抗病性等多种生理功能(Rong et al., 2016)。以拟南芥为例, AtCAD1、AtCAD4和AtCAD5参与木质素合成(Sibout et al., 2003), 而AtCAD7和AtCAD8与植物抗性有关(Kim et al., 2004; Barakat et al., 2009)。AtCAD2和AtCAD3也显示出对羟基肉桂醛的活性, 但是对木质素沉积几乎无影响, 主要参与生物/非生物胁迫反应(Eudes et al., 2006)。AmCAD重组蛋白表现出较强的pH敏感性和较高的温度稳定性。在以松柏醛为底物时, 25-55°C之间AmCAD蛋白均能维持较高的活力。AmCAD催化不同底物的最适pH均约为7, 环境pH偏酸或偏碱都会导致重组蛋白的催化活力迅速降低。已报道不同物种中参与木质素合成的CAD均表现出相似的pH敏感性(Bukh et al., 2012)。由此可见, 植物细胞质中pH值的稳定性对于木质素单体的高效合成具有重要作用。结构的保守性和定位的多样性均表明, 蒙古冰草中AmCAD蛋白可能在体外具有相似的底物催化能力, 但在胞内则发挥不同的功能, 从而影响蒙古冰草的生长、抗逆以及品质等多个方面。
尽管CAD基因家族成员众多, 但参与木质素合成的关键基因仅占其中的一小部分。对蒙古冰草AmCAD进行系统进化分析, 结果表明这些蛋白可分为4个亚家族, 其中AmCAD与其它参与木质素合成的CAD一起聚类于Group I中。结构分析表明, AmCAD能够形成同型二聚体, 与拟南芥AtCAD5 (Youn et al., 2006)和二穗短柄草BdCAD5 (Tobias and Chow, 2005)一致。生物信息学分析结果也表明, AmCAD可能作为AtCAD5的同源蛋白参与木质素合成过程。对AmCAD重组蛋白底物催化活性的分析进一步证实了这一结论。与其它物种中的CAD蛋白相比, AmCAD具有更强的底物催化活性。在小麦中, TaCAD1对松柏醛、芥子醛和对香豆醛的Km值分别为21.94、40.63和27.16 μmol∙L-1 (Ma, 2010)。在青蒿(Artemisia apiacea)中, AaCAD对芥子醛和松柏醛的Km值分别为15.61和10.83 μmol∙L-1 (Li et al., 2012)。在银合欢(Leucaena leucocephala)中, LlCAD对芥子醛的Km值为23.8 μmol∙L-1, 对松柏醛的Km为18.1 μmol∙L-1 (Pandey et al., 2011)。研究表明, 参与木质素合成的CAD基因主要在茎秆中表达。小麦TaCAD1呈现出独特的器官特异性表达模式, 在茎中表达量高, 在叶中表达量非常低, 在根中检测不到表达量(Ma, 2010)。Tsuruta等(2007)在高粱茎中也发现了CAD基因的高表达, 该基因参与次生细胞壁中木质素含量和组分调控。对蒙古冰草不同组织中AmCAD表达量的分析显示该基因在茎秆中特异性表达。结合生物信息学、体外酶活以及组织特异性表达分析结果, 表明AmCAD是蒙古冰草中参与木质素合成的重要基因。
研究表明, CAD蛋白的底物偏好性与木质素的结构组成相关(Bukh et al., 2012)。木质素主要包括对羟苯基木质素(H型木质素)、愈创木基木质素(G型木质素)和紫丁香基木质素(S型木质素)等结构单元, 单子叶植物的木质素主要由G型和S型木质素构成(Zeng et al., 2013)。酶活测定和酶动力学分析结果表明, AmCAD重组蛋白对松柏醛、芥子醛和对香豆醛等不同肉桂醛底物均具有很强的催化能力, 同时也表现出对松柏醛和芥子醛的偏好性。在最适反应条件下, 该蛋白对松柏醛和芥子醛亲和力较强, Km值分别为8.94和10.35 μmol∙L-1。AmCAD蛋白对3种底物的催化能力和亲和力强弱与木质素中H型、G型和S型含量相对应, 表明AmCAD的底物偏好性可能影响蒙古冰草木质素结构和各类型单体的组分。
通过对CAD基因进行调控, 研究人员能够定向改变木质素含量或木质素单体比例, 进而提高能源作物的酶解糖化效率或牧草作物的消化率。玉米(Zea mays)和高粱的棕色中脉突变体表现出叶中脉和茎的红棕色色素沉积, 这种表型通常与木质素含量降低和组成改变及青贮饲料消化率提高有关(Cherney et al., 1991; Vermerris et al., 2002)。玉米bm1突变体中CAD基因的碱基突变影响CAD蛋白的催化活性, 降低了该突变体细胞壁中的木质素含量以及G型和S型单体含量(Halpin et al., 1998)。高粱bmr6突变体中CAD基因的突变也导致木质素含量和组成发生改变, 突变体植株的生物质具有更高的糖化效率(Sattler et al., 2009)。在柳枝稷(Panicum virgatum)中, 研究人员利用RNAi技术抑制PvCAD基因的表达, 获得了木质素含量降低的基因工程植株, 显著提高了柳枝稷的酶解糖化效率和牧草消化率(Fu et al., 2011)。我国缺乏高品质牧草品种, AmCAD基因的筛选和鉴定为蒙古冰草品质改良和种质创新提供了重要的基因资源。
此外, CAD基因也在植物的抗逆性和抗病性等过程中发挥关键作用(Park et al., 2018; Lee et al., 2021)。已有研究表明, 在生物和非生物胁迫下甘薯(Dioscorea esculenta)和银杏(Ginkgo biloba) CAD1基因被强烈诱导表达(Saballos et al., 2009; Xu et al., 2011; Pan et al., 2014)。在棉花中也发现, 几种GhCAD在大丽弧菌感染时被诱导表达(Xiao et al., 2021)。对毛竹(Phyllostachys edulis) PheCAD基因响应非生物胁迫的研究发现, 经过干旱、ABA和盐等不同非生物胁迫处理后, PheCAD2均被显著诱导表达(Vasupalli et al., 2021)。在蒙古冰草中, AmCAD基因的表达量受干旱胁迫强烈诱导, 表明该基因既参与木质素合成, 也可能参与对非生物胁迫的防御(Li et al., 2001)。
本研究从蒙古冰草中筛选和鉴定了参与木质素合成的关键基因AmCAD, 其编码蛋白具有很强的催化肉桂醛类底物反应的能力, 参与木质素合成的同时也显著响应干旱胁迫。因此, 该基因可能参与木质素等重要苯丙素类化合物的合成, 同时与蒙古冰草的耐旱性相关。该基因作为蒙古冰草分子改良的候选基因, 对于提高蒙古冰草牧草品质和抗逆性具有重要意义。然而, 目前蒙古冰草遗传转化体系的欠缺严重限制了这一重要本土牧草的分子育种工作。本文作者研究团队正致力于突破蒙古冰草的遗传转化瓶颈, 在包括AmCAD的基因功能鉴定的基础上, 通过基因编辑等技术手段, 继续推进蒙古冰草的分子育种工作。
作者贡献声明
王贺萍: 撰写论文; 孙震、刘雨辰和苏彦龙: 分析数据并提供技术支持; 杜锦瑜: 提供实验协助; 赵彦、赵竑博、王召明、苑峰、刘亚玲和吴振映: 提供实验思路; 何峰和付春祥: 设计实验并修改文章。
参考文献

Lignin is a phenolic heteropolymer in secondary cell walls that plays a major role in the development of plants and their defense against pathogens. The biosynthesis of monolignols, which represent the main component of lignin involves many enzymes. The cinnamyl alcohol dehydrogenase (CAD) is a key enzyme in lignin biosynthesis as it catalyzes the final step in the synthesis of monolignols. The CAD gene family has been studied in Arabidopsis thaliana, Oryza sativa and partially in Populus. This is the first comprehensive study on the CAD gene family in woody plants including genome organization, gene structure, phylogeny across land plant lineages, and expression profiling in Populus.The phylogenetic analyses showed that CAD genes fall into three main classes (clades), one of which is represented by CAD sequences from gymnosperms and angiosperms. The other two clades are represented by sequences only from angiosperms. All Populus CAD genes, except PoptrCAD 4 are distributed in Class II and Class III. CAD genes associated with xylem development (PoptrCAD 4 and PoptrCAD 10) belong to Class I and Class II. Most of the CAD genes are physically distributed on duplicated blocks and are still in conserved locations on the homeologous duplicated blocks. Promoter analysis of CAD genes revealed several motifs involved in gene expression modulation under various biological and physiological processes. The CAD genes showed different expression patterns in poplar with only two genes preferentially expressed in xylem tissues during lignin biosynthesis.The phylogeny of CAD genes suggests that the radiation of this gene family may have occurred in the early ancestry of angiosperms. Gene distribution on the chromosomes of Populus showed that both large scale and tandem duplications contributed significantly to the CAD gene family expansion. The duplication of several CAD genes seems to be associated with a genome duplication event that happened in the ancestor of Salicaceae. Phylogenetic analyses associated with expression profiling and results from previous studies suggest that CAD genes involved in wood development belong to Class I and Class II. The other CAD genes from Class II and Class III may function in plant tissues under biotic stresses. The conservation of most duplicated CAD genes, the differential distribution of motifs in their promoter regions, and the divergence of their expression profiles in various tissues of Populus plants indicate that genes in the CAD family have evolved tissue-specialized expression profiles and may have divergent functions.

Cinnamyl alcohol dehydrogenase (CAD) catalyses the final step of the monolignol biosynthesis, the conversion of cinnamyl aldehydes to alcohols, using NADPH as a cofactor. Seven members of the CAD gene family were identified in the genome of Brachypodium distachyon and five of these were isolated and cloned from genomic DNA. Semi-quantitative reverse-transcription PCR revealed differential expression of the cloned genes, with BdCAD5 being expressed in all tissues and highest in root and stem while BdCAD3 was only expressed in stem and spikes. A phylogenetic analysis of CAD-like proteins placed BdCAD5 on the same branch as bona fide CAD proteins from maize (ZmCAD2), rice (OsCAD2), sorghum (SbCAD2) and Arabidopsis (AtCAD4, 5). The predicted three-dimensional structures of both BdCAD3 and BdCAD5 resemble that of AtCAD5. However, the amino-acid residues in the substrate-binding domains of BdCAD3 and BdCAD5 are distributed symmetrically and BdCAD3 is similar to that of poplar sinapyl alcohol dehydrogenase (PotSAD). BdCAD3 and BdCAD5 expressed and purified from Escherichia coli both showed a temperature optimum of about 50 °C and molar weight of 49 kDa. The optimal pH for the reduction of coniferyl aldehyde were pH 5.2 and 6.2 and the pH for the oxidation of coniferyl alcohol were pH 8 and 9.5, for BdCAD3 and BdCAD5 respectively. Kinetic parameters for conversion of coniferyl aldehyde and coniferyl alcohol showed that BdCAD5 was clearly the most efficient enzyme of the two. These data suggest that BdCAD5 is the main CAD enzyme for lignin biosynthesis and that BdCAD3 has a different role in Brachypodium. All CAD enzymes are cytosolic except for BdCAD4, which has a putative chloroplast signal peptide adding to the diversity of CAD functions.

The cinnamyl alcohol dehydrogenase (AtCAD) multigene family in Arabidopsis is composed of nine genes. Our previous studies focused on the two isoforms AtCAD C and AtCAD D which show a high homology to those related to lignification in other plants. This study focuses on the seven other Arabidopsis CAD for which functions are not yet elucidated. Their expression patterns were determined in different parts of Arabidopsis. Only CAD 1 protein can be detected in elongating stems, flowers, and siliques using Western-blot analysis. Tissue specific expression of CAD 1, B1, and G genes was determined using their promoters fused to the GUS reporter gene. CAD 1 expression was observed in primary xylem in accordance with a potential role in lignification. Arabidopsis T-DNA mutants knockout for the different genes CAD genes were characterized. Their stems displayed no substantial reduction of CAD activities for coniferyl and sinapyl alcohols as well as no modifications of lignin quantity and structure in mature inflorescence stems. Only a small reduction of lignin content could be observed in elongating stems of Atcad 1 mutant. These CAD genes in combination with the CAD D promoter were used to complement a CAD double mutant severely altered in lignification (cad c cad d). The expression of AtCAD A, B1, B2, F, and G had no effect on restoring a normal lignin profile of this mutant. In contrast, CAD 1 complemented partly this mutant as revealed by the partial restoration of conventional lignin units and by the decrease in the frequency of beta-O-4 linked p-OH cinnamaldehydes.

NAC转录因子具有调控植物生长发育及应对非生物逆境胁迫等多种功能。为筛选蒙古冰草(Agropyron mongolicum Keng)抗旱相关的NAC转录因子,本研究基于转录组测序数据,利用生物信息学方法对蒙古冰草NAC转录因子的结构、理化性质、进化关系和保守性等进行分析,同时运用实时荧光定量技术对抗旱相关的2个NAC基因进行差异表达分析。结果表明:筛选出26个蒙古冰草NAC家族成员,其中15个具有完整NAC转录因子结构域,其均属于亲水性蛋白,大部分定位于细胞核;15个蒙古冰草NAC转录因子的蛋白磷酸化位点均含有丝氨酸、苏氨酸和酪氨酸。进化关系比对显示AmNAC100和AmNAC102-2转录因子与已知有抗旱功能的NAC蛋白同源性极高,其二、三级结构主要以无规则卷曲为主,motif1,2,3,8为其共有的保守基序,多序列比对将保守结构域划分为子域A—E。差异表达分析发现AmNAC100和AmNAC102-2基因相对表达量均高于对照(CK),在蒙古冰草干旱胁迫中起正调控作用。

Brown-midrib (bm) mutants of maize have modified lignin of reddish-brown colour. Although four independent bm loci are known, only one of the mutant genes has been previously identified. We report here that maize bm1, one of the less characterised mutants, shows severely reduced CAD activity in lignified tissues, resulting in the production of a modified lignin. Both the total lignin content and the structure of the polymer are altered by the mutation. We further describe the isolation and characterisation of the maize CAD cDNA and mapping of the CAD gene. CAD maps very closely to the known location of bm1 and co-segregates with the bm1 locus in two independent recombinant inbred populations. These data strongly support the premise that maize bm1 directly affects expression of the CAD gene.

Stress signalling and regulatory networks controlling expression of target genes are the basis of plant response to drought. Roots are the first organs exposed to water deficiency in the soil and are the place of drought sensing. Signalling cascades transfer chemical signals toward the shoot and initiate molecular responses that lead to the biochemical and morphological changes that allow plants to be protected against water loss and to tolerate stress conditions. Here, we present an overview of signalling network and gene expression regulation pathways that are actively induced in roots under drought stress. In particular, the role of several transcription factor (TF) families, including DREB, AP2/ERF, NAC, bZIP, MYC, CAMTA, Alfin-like and Q-type ZFP, in the regulation of root response to drought are highlighted. The information provided includes available data on mutual interactions between these TFs together with their regulation by plant hormones and other signalling molecules. The most significant downstream target genes and molecular processes that are controlled by the regulatory factors are given. These data are also coupled with information about the influence of the described regulatory networks on root traits and root development which may translate to enhanced drought tolerance. This is the first literature survey demonstrating the gene expression regulatory machinery that is induced by drought stress, presented from the perspective of roots. © The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology. All rights reserved. For permissions, please email: journals.permissions@oup.com.

Cinnamyl alcohol dehydrogenase (CAD; EC 1.1.1.195) has been thought to mediate the reduction of both coniferaldehyde and sinapaldehyde into guaiacyl and syringyl monolignols in angiosperms. Here, we report the isolation of a novel aspen gene (PtSAD) encoding sinapyl alcohol dehydrogenase (SAD), which is phylogenetically distinct from aspen CAD (PtCAD). Liquid chromatography-mass spectrometry-based enzyme functional analysis and substrate level-controlled enzyme kinetics consistently demonstrated that PtSAD is sinapaldehyde specific and that PtCAD is coniferaldehyde specific. The enzymatic efficiency of PtSAD for sinapaldehyde was approximately 60 times greater than that of PtCAD. These data suggest that in addition to CAD, discrete SAD function is essential to the biosynthesis of syringyl monolignol in angiosperms. In aspen stem primary tissues, PtCAD was immunolocalized exclusively to xylem elements in which only guaiacyl lignin was deposited, whereas PtSAD was abundant in syringyl lignin-enriched phloem fiber cells. In the developing secondary stem xylem, PtCAD was most conspicuous in guaiacyl lignin-enriched vessels, but PtSAD was nearly absent from these elements and was conspicuous in fiber cells. In the context of additional protein immunolocalization and lignin histochemistry, these results suggest that the distinct CAD and SAD functions are linked spatiotemporally to the differential biosynthesis of guaiacyl and syringyl lignins in different cell types. SAD is required for the biosynthesis of syringyl lignin in angiosperms.

植物木质素生物合成调控研究已在造纸树种与饲草品质的改良中取得了许多进展。随着对木质纤维原料乙醇发酵研究的兴起, 植物木质素合成调控再次成为研究热点。该文总结了目前生物质能源利用的现状, 同时针对木质素在木质纤维乙醇发酵中的限制作用, 综述了近年来植物木质素合成调控的研究进展, 提出了今后的研究方向和内容, 并展望了木质素合成调控在木质纤维乙醇发酵中的应用。

饲草种质资源是国家基础性和战略性资源, 事关草种业振兴全局, 是现代农牧业发展和生态保护修复的基础。该文综述了全球饲草种质资源的种类、分布、特征以及保存状况, 分析了目前存在的问题并提出建议, 旨在更好地保护和利用我国饲草种质资源。

CmCAD2 and CmCAD3 function more positively than CmCAD1 in oriental melon for lignin synthesis which is important to ensure internal water status and thus for drought tolerance. Well-lignification may be the guarantee of efficient axial water transport and barrier of lateral water flow in oriental melon tolerating drought stress, however remains to be verified. As an important enzyme in monolignol synthesis pathway, five cinnamyl alcohol dehydrogenase (CAD) genes were generally induced in melon seedlings by drought. Here we further revealed the roles of CmCAD1, 2, and 3 in lignin synthesis and for drought tolerance. Results found that overexpressing CmCAD2 or 3 strongly recovered CAD activities, lignin synthesis and composition in Arabidopsis cadc cadd, whose lignin synthesis is disrupted, while CmCAD1 functioned modestly. In melon seedlings, silenced CmCAD2 and 3 individually or collectively decreased CAD activities and lignin depositions drastically, resulting in dwarfed phenotypes. Reduced lignin, mainly composed by guaiacyl units catalyzed by CmCAD3, is mainly due to the limited lignification in tracheary elements and development of Casparion strip. While CmCAD1 and 2 exhibited catalysis to p-coumaraldehyde and sinapaldehyde, respectively. Compared with CmCAD1, drought treatments revealed higher sensitivity of CmCAD2 and/or 3 silenced melon seedlings, accompanying with lower relative water contents, water potentials and relatively higher total soluble sugar contents. Slightly up-regulated expressions of aquaporin genes together with limited lignification might imply higher lateral water loss in stems of silenced lines. In Arabidopsis, CmCAD2 and 3 transgenic lines enhanced cadc cadd drought tolerance through recovering lignin synthesis and root development, accompanying with decreased electrolyte leakage ratios and increased RWCs, thus improved survival rates. Briefly, lignin synthesized by CmCAD2 and 3 functions importantly for drought tolerance in melon.

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.Copyright 2001 Elsevier Science (USA).

The plant aromatic alcohol dehydrogenase, cinnamyl alcohol dehydrogenase (CAD2 from Eucalyptus) was found by sequence analysis of its cloned gene to be homologous to a range of dehydrogenases including alcohol dehydrogenases, L-threonine-3-dehydrogenase, D-xylose reductase and sorbitol dehydrogenase. A homology model of CAD2 was built using the X-ray crystallographic coordinates of horse-liver alcohol dehydrogenase to provide the template, with additional modelling input from other analogous regions of structure from similar enzymes where necessary. The structural model thus produced rationalised the Zn-binding properties of CAD2, indicated the possession of a Rossmann fold (GXGXXG motif), and explained the class A stereospecificity (pro-R hydrogen removal from substrate alcohol) and aromatic substrate specificity of the enzyme. A range of potential ligands was designed based on the homology model and tested as inhibitors of CAD2 and horse liver alcohol dehydrogenase.

The enzymes cinnamoyl-CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) catalyze the two key reduction reactions in the conversion of cinnamic acid derivatives into monolignol building blocks for lignin polymers in plant cell walls. Here, we describe detailed functional and structural analyses of CCRs from Medicago truncatula and Petunia hybrida and of an atypical CAD (CAD2) from M. truncatula. These enzymes are closely related members of the short-chain dehydrogenase/reductase (SDR) superfamily. Our structural studies support a reaction mechanism involving a canonical SDR catalytic triad in both CCR and CAD2 and an important role for an auxiliary cysteine unique to CCR. Site-directed mutants of CAD2 (Phe226Ala and Tyr136Phe) that enlarge the phenolic binding site result in a 4- to 10-fold increase in activity with sinapaldehyde, which in comparison to the smaller coumaraldehyde and coniferaldehyde substrates is disfavored by wild-type CAD2. This finding demonstrates the potential exploitation of rationally engineered forms of CCR and CAD2 for the targeted modification of monolignol composition in transgenic plants. Thermal denaturation measurements and structural comparisons of various liganded and unliganded forms of CCR and CAD2 highlight substantial conformational flexibility of these SDR enzymes, which plays an important role in the establishment of catalytically productive complexes of the enzymes with their NADPH and phenolic substrates.

Cinnamyl alcohol dehydrogenase (CAD) is involved in the final step of the phenylpropanod pathway, catalyzing the NADPH-dependent reduction of hydroxy-cinnamaldehydes into the corresponding alcohols. The rice genome contains twelve CAD and CAD-like genes, collectively called OsCADs. To elucidate the biochemical function of the OsCADs, OsCAD1, 2, 6, and 7, which are highly expressed in rice, were cloned from rice tissues. The cloned OsCADs were heterologously expressed in Escherichia coli as His-tag fusion proteins. The activity assay of the recombinant OsCADs showed that OsCAD2, 6, and 7 have CAD activity toward hydroxycinnamaldehydes, but OsCAD1 has no detectable catalytic activity. The kinetic parameters of the enzyme reactions demonstrated that OsCAD2 has the highest catalytic activity among the examined enzymes. This result agrees well with the finding that the Zn binding and NADPH binding motifs and the residues constituting the substrate binding pocket in bona fide plant CADs were fully conserved in OsCAD2. Although they have large variations in the residue for the substrate binding pocket, OsCAD6 and 7 catalyzed the reduction of hydroxycinnamaldehydes with a similar efficiency. Alignment of amino acid sequences showed that OsCAD1 lacks the GxxxxP motif for NADPH binding and has mismatches in residues important in the reduction process, which could be responsible for the loss of catalytic activity. OsCAD2 belongs to CAD Class I with bona fide CADs from other plant species and is constitutively expressed throughout the developmental stages of rice, with preferential expression in actively lignifying tissues such as the root, stem, and panicle, suggesting that it is mainly involved in developmental lignification in rice. The expression of OsCAD2 was also induced by biotic and abiotic stresses such as Xanthomonas oryzae pv. oryzae (Xoo) infection and UV-irradiation, suggesting that it plays a role in the defense response of rice, in addition to a bona fide role in developmental lignification. OsCAD6 and 7 belong in CAD Class II. Their expression is relatively lower than that of OsCAD2 and is confined to certain tissues, such as the leaf sheath, stem, and panicle. The expression of OsCAD6 was stimulated by Xoo infection and UV-irradiation. Thus OsCAD6 appears to be an inducible OsCAD that is likely involved in the defense response of rice against biotic and abiotic stresses.

The objective of this study was to identify barley leaf proteins differentially regulated in response to drought and heat and the combined stresses in context of the morphological and physiological changes that also occur. The Syrian landrace Arta and the Australian cultivar Keel were subjected to drought, high temperature, or a combination of both treatments starting at heading. Changes in the leaf proteome were identified using differential gel electrophoresis and mass spectrometry. The drought treatment caused strong reductions of biomass and yield, while photosynthetic performance and the proteome were not significantly changed. In contrast, the heat treatment and the combination of heat and drought reduced photosynthetic performance and caused changes of the leaf proteome. The proteomic analysis identified 99 protein spots differentially regulated in response to heat treatment, 14 of which were regulated in a genotype-specific manner. Differentially regulated proteins predominantly had functions in photosynthesis, but also in detoxification, energy metabolism, and protein biosynthesis. The analysis indicated that de novo protein biosynthesis, protein quality control mediated by chaperones and proteases, and the use of alternative energy resources, i.e. glycolysis, play important roles in adaptation to heat stress. In addition, genetic variation identified in the proteome, in plant growth and photosynthetic performance in response to drought and heat represent stress adaption mechanisms to be exploited in future crop breeding efforts.

The content and composition of the plant cell wall polymer lignin affect plant fitness, carbon sequestration potential, and agro-industrial processing. These characteristics, are heavily influenced by the supply of hydroxycinnamyl alcohol precursors synthesized by the enzyme cinnamyl alcohol dehydrogenase (CAD). In angiosperms, CAD is encoded by a multigene family consisting of members thought to have distinct roles in different stages of plant development. Due to the high sequence similarity among CAD genes, it has been challenging to identify and study the role of the individual genes without a genome sequence. Analysis of the recently released sorghum genome revealed the existence of 14 CAD-like genes at seven genomic locations. Comparisons with maize and rice revealed subtle differences in gene number, arrangement, and expression patterns. Sorghum CAD2 is the predominant CAD involved in lignification based on the phylogenetic relationship with CADs from other species and genetic evidence showing that a set of three allelic brown midrib (bmr) lignin mutants contained mutations in this gene. The impact of the mutations on the structure of the protein was assessed using molecular modeling based on X-ray crystallography data of the closely related Arabidopsis CAD5. The modeling revealed unique changes in structure consistent with the observed phenotypes of the mutants.

Brown midrib6 (bmr6) affects phenylpropanoid metabolism, resulting in reduced lignin concentrations and altered lignin composition in sorghum (Sorghum bicolor). Recently, bmr6 plants were shown to have limited cinnamyl alcohol dehydrogenase activity (CAD; EC 1.1.1.195), the enzyme that catalyzes the conversion of hydroxycinnamoyl aldehydes (monolignals) to monolignols. A candidate gene approach was taken to identify Bmr6. Two CAD genes (Sb02g024190 and Sb04g005950) were identified in the sorghum genome based on similarity to known CAD genes and through DNA sequencing a nonsense mutation was discovered in Sb04g005950 that results in a truncated protein lacking the NADPH-binding and C-terminal catalytic domains. Immunoblotting confirmed that the Bmr6 protein was absent in protein extracts from bmr6 plants. Phylogenetic analysis indicated that Bmr6 is a member of an evolutionarily conserved group of CAD proteins, which function in lignin biosynthesis. In addition, Bmr6 is distinct from the other CAD-like proteins in sorghum, including SbCAD4 (Sb02g024190). Although both Bmr6 and SbCAD4 are expressed in sorghum internodes, an examination of enzymatic activity of recombinant Bmr6 and SbCAD4 showed that Bmr6 had 1 to 2 orders of magnitude greater activity for monolignol substrates. Modeling of Bmr6 and SbCAD4 protein structures showed differences in the amino acid composition of the active site that could explain the difference in enzyme activity. These differences include His-57, which is unique to Bmr6 and other grass CADs. In summary, Bmr6 encodes the major CAD protein involved in lignin synthesis in sorghum, and the bmr6 mutant is a null allele.

Barley is a major cereal crop for temperate climates, and a diploid genetic model for polyploid wheat. Cereal straw biomass is an attractive source of feedstock for green technologies but lignin, a key determinant of feedstock recalcitrance, complicates bio-conversion processes. However, manipulating lignin content to improve the conversion process could negatively affect agronomic traits. An alternative approach is to manipulate lignin composition which influences the physical and chemical properties of straw. This study validates the function of a barley ferulate 5-hydroxylase gene and demonstrates that its downregulation using the RNA-interference approach substantially impacts lignin composition. We identified five barley genes having putative ferulate 5-hydroxylase activity. Downregulation of HvF5H1 substantially reduced the lignin syringyl/guaiacyl (S/G) ratio in straw while the lignin content, straw mechanical properties, plant growth habit, and grain characteristics all remained unaffected. Metabolic profiling revealed significant changes in the abundance of 173 features in the HvF5H1-RNAi lines. The drastic changes in the lignin polymer of transgenic lines highlight the plasticity of barley lignification processes and the associated potential for manipulating and improving lignocellulosic biomass as a feedstock for green technologies. On the other hand, our results highlight some differences between the lignin biosynthetic pathway in barley, a temperate climate grass, and the warm climate grass, rice, and underscore potential diversity in the lignin biosynthetic pathways in grasses.

Studying Arabidopsis mutants of the phenylpropanoid pathway has unraveled several biosynthetic steps of monolignol synthesis. Most of the genes leading to monolignol synthesis have been characterized recently in this herbaceous plant, except those encoding cinnamyl alcohol dehydrogenase (CAD). We have used the complete sequencing of the Arabidopsis genome to highlight a new view of the complete CAD gene family. Among nine AtCAD genes, we have identified the two distinct paralogs AtCAD-C and AtCAD-D, which share 75% identity and are likely to be involved in lignin biosynthesis in other plants. Northern, semiquantitative restriction fragment-length polymorphism-reverse transcriptase-polymerase chain reaction and western analysis revealed that AtCAD-C and AtCAD-D mRNA and protein ratios were organ dependent. Promoter activities of both genes are high in fibers and in xylem bundles. However, AtCAD-C displayed a larger range of sites of expression than AtCAD-D. Arabidopsis null mutants (Atcad-D and Atcad-C) corresponding to both genes were isolated. CAD activities were drastically reduced in both mutants, with a higher impact on sinapyl alcohol dehydrogenase activity (6% and 38% of residual sinapyl alcohol dehydrogenase activities for Atcad-D and Atcad-C, respectively). Only Atcad-D showed a slight reduction in Klason lignin content and displayed modifications of lignin structure with a significant reduced proportion of conventional S lignin units in both stems and roots, together with the incorporation of sinapaldehyde structures ether linked at Cbeta. These results argue for a substantial role of AtCAD-D in lignification, and more specifically in the biosynthesis of sinapyl alcohol, the precursor of S lignin units.

Lignin biosynthesis enzymes form complexes for metabolic channelling during lignification and these enzymes also play an essential role in biotic and abiotic stress response. Cinnamyl alcohol dehydrogenase (CAD) is a vital enzyme that catalyses the reduction of aldehydes to alcohols, which is the final step in the lignin biosynthesis pathway. In the present study, we identified 49 CAD enzymes in five Bambusoideae species and analysed their phylogenetic relationships and conserved domains. Expression analysis of Moso bamboo PheCAD genes in several developmental tissues and stages revealed that among the PheCAD genes, PheCAD2 has the highest expression level and is expressed in many tissues and PheCAD1, PheCAD6, PheCAD8 and PheCAD12 were also expressed in most of the tissues studied. Co-expression analysis identified that the PheCAD2 positively correlates with most lignin biosynthesis enzymes, indicating that PheCAD2 might be the key enzyme involved in lignin biosynthesis. Further, more than 35% of the co-expressed genes with PheCADs were involved in biotic or abiotic stress responses. Abiotic stress transcriptomic data (SA, ABA, drought, and salt) analysis identified that PheCAD2, PheCAD3 and PheCAD5 genes were highly upregulated, confirming their involvement in abiotic stress response. Through yeast two-hybrid analysis, we found that PheCAD1, PheCAD2 and PheCAD8 form homo-dimers. Interestingly, BiFC and pull-down experiments identified that these enzymes form both homo- and hetero- dimers. These data suggest that PheCAD genes are involved in abiotic stress response and PheCAD2 might be a key lignin biosynthesis pathway enzyme. Moreover, this is the first report to show that three PheCAD enzymes form complexes and that the formation of PheCAD homo- and hetero- dimers might be tissue specific.

The four brown midrib (bm) mutants of maize have a reduced content and altered subunit composition of the cell wall polymer lignin. The bm mutations have traditionally been considered completely recessive, because the brown midrib phenotype is only apparent in plants homozygous for the mutation. In addition to an effect on cell wall composition, some bm mutations have been shown to affect flowering time. We had preliminary evidence for a dosage effect of the Bm1 locus on flowering time, which prompted this detailed study on the Bm1 locus. In this study, near-isogenic lines (in an A619 background) with zero, one or two bm1 mutant alleles were compared. The bm1 heterozygotes flowered significantly earlier than both the wild-type plants and bm1 mutants. This difference can at least be partly attributed to an accelerated growth rate in the later stages of plant development. Furthermore, Fourier transform infrared spectroscopy revealed that the cell wall composition of the bm1 heterozygous plants is distinct from both the bm1 and wild-type homozygotes. The combination of the data on flowering time and the data on cell wall composition provide evidence for a dosage effect at the Bm1 locus.

用不同浓度的NaCl溶液处理蒙古冰草幼苗后,测定了不同时间胁迫下植株地上部分鲜重和离子含量。结果表明:轻度盐胁迫对蒙古冰草幼苗的生长有一定的促进作用,而中度和重度盐胁迫对幼苗生长有明显的抑制作用。经分析认为,轻度盐胁迫下蒙古冰草可以将Na<sup>+</sup>等有害离子截流在根部,抑制其向地上部分的运输,而中度和重度盐胁迫下过多的Na<sup>+</sup>等有害离子运输到地上代谢活跃部分,限制了对K<sup>+</sup>、Ca<sup>2+</sup>等的吸收,造成植株的营养亏缺,从而引起植株的伤害。

GhMYB4 acts as a negative regulator in lignin biosynthesis, which results in alteration of cell wall integrity and activation of cotton defense response. Verticillium wilt of cotton (Gossypium hirsutum) caused by the soil-borne fungus Verticillium dahliae (V. dahliae) represents one of the most important constraints of cotton production worldwide. Mining of the genes involved in disease resistance and illuminating the molecular mechanisms that underlie this resistance is of great importance in cotton breeding programs. Defense-induced lignification in plants is necessary for innate immunity, and there are reports of a correlation between increased lignification and disease resistance. In this study, we present an example in cotton whereby plants with reduced lignin content also exhibit enhanced disease resistance. We identified a negative regulator of lignin synthesis, in cotton encoded in GhMYB4. Overexpression of GhMYB4 in cotton and Arabidopsis enhanced resistance to V. dahliae with reduced lignin deposition. Moreover, GhMYB4 could bind the promoters of several genes involved in lignin synthesis, such as GhC4H-1, GhC4H-2, Gh4CL-4, and GhCAD-3, and impair their expression. The reduction of lignin content in GhMYB4-overexpressing cotton led to alterations of cell wall integrity (CWI) and released more oligogalacturonides (OGs) which may act as damage-associated molecular patterns (DAMPs) to stimulate plant defense responses. In support of this hypothesis, exogenous application with polygalacturonic acid (PGA) in cotton activated biosynthesis of jasmonic acid (JA) and JA-mediated defense against V. dahliae, similar to that described for cotton plants overexpressing GhMYB4. This study provides a new candidate gene for cotton disease-resistant breeding and an increased understanding of the relationship between lignin synthesis, OG release, and plant immunity.

The incompatible pathosystem between resistant cotton (Gossypium barbadense cv. 7124) and Verticillium dahliae strain V991 was used to study the cotton transcriptome changes after pathogen inoculation by RNA-Seq. Of 32,774 genes detected by mapping the tags to assembly cotton contigs, 3442 defence-responsive genes were identified. Gene cluster analyses and functional assignments of differentially expressed genes indicated a significant transcriptional complexity. Quantitative real-time PCR (qPCR) was performed on selected genes with different expression levels and functional assignments to demonstrate the utility of RNA-Seq for gene expression profiles during the cotton defence response. Detailed elucidation of responses of leucine-rich repeat receptor-like kinases (LRR-RLKs), phytohormone signalling-related genes, and transcription factors described the interplay of signals that allowed the plant to fine-tune defence responses. On the basis of global gene regulation of phenylpropanoid metabolism-related genes, phenylpropanoid metabolism was deduced to be involved in the cotton defence response. A closer look at the expression of these genes, enzyme activity, and lignin levels revealed differences between resistant and susceptible cotton plants. Both types of plants showed an increased level of expression of lignin synthesis-related genes and increased phenylalanine-ammonia lyase (PAL) and peroxidase (POD) enzyme activity after inoculation with V. dahliae, but the increase was greater and faster in the resistant line. Histochemical analysis of lignin revealed that the resistant cotton not only retains its vascular structure, but also accumulates high levels of lignin. Furthermore, quantitative analysis demonstrated increased lignification and cross-linking of lignin in resistant cotton stems. Overall, a critical role for lignin was believed to contribute to the resistance of cotton to disease.

The cinnamyl alcohol dehydrogenase (CAD) multigene family in planta encodes proteins catalyzing the reductions of various phenylpropenyl aldehyde derivatives in a substrate versatile manner, and whose metabolic products are the precursors of structural lignins, health-related lignans, and various other metabolites. In Arabidopsis thaliana, the two isoforms, AtCAD5 and AtCAD4, are the catalytically most active being viewed as mainly involved in the formation of guaiacyl/syringyl lignins. In this study, we determined the crystal structures of AtCAD5 in the apo-form and as a binary complex with NADP+, respectively, and modeled that of AtCAD4. Both AtCAD5 and AtCAD4 are dimers with two zinc ions per subunit and belong to the Zn-dependent medium chain dehydrogenase/reductase (MDR) superfamily, on the basis of their overall 2-domain structures and distribution of secondary structural elements. The catalytic Zn2+ ions in both enzymes are tetrahedrally coordinated, but differ from those in horse liver alcohol dehydrogenase since the carboxyl side-chain of Glu70 is ligated to Zn2+ instead of water. Using AtCAD5, site-directed mutagenesis of Glu70 to alanine resulted in loss of catalytic activity, thereby indicating that perturbation of the Zn2+ coordination was sufficient to abolish catalytic activity. The substrate-binding pockets of both AtCAD5 and AtCAD4 were also examined, and found to be significantly different and smaller compared to that of a putative aspen sinapyl alcohol dehydrogenase (SAD) and a putative yeast CAD. While the physiological roles of the aspen SAD and the yeast CAD are uncertain, they nevertheless have a high similarity in the overall 3D structures to AtCAD5 and 4. With the bona fide CAD's from various species, nine out of the twelve residues which constitute the proposed substrate-binding pocket were, however, conserved. This is provisionally considered as indicative of a characteristic fingerprint for the CAD family.

Lignin content and composition are two important agronomic traits for the utilization of agricultural residues. Rice (Oryza sativa) gold hull and internode phenotype is a classical morphological marker trait that has long been applied to breeding and genetics study. In this study, we have cloned the GOLD HULL AND INTERNODE2 (GH2) gene in rice using a map-based cloning approach. The result shows that the gh2 mutant is a lignin-deficient mutant, and GH2 encodes a cinnamyl-alcohol dehydrogenase (CAD). Consistent with this finding, extracts from roots, internodes, hulls, and panicles of the gh2 plants exhibited drastically reduced CAD activity and undetectable sinapyl alcohol dehydrogenase activity. When expressed in Escherichia coli, purified recombinant GH2 was found to exhibit strong catalytic ability toward coniferaldehyde and sinapaldehyde, while the mutant protein gh2 completely lost the corresponding CAD and sinapyl alcohol dehydrogenase activities. Further phenotypic analysis of the gh2 mutant plants revealed that the p-hydroxyphenyl, guaiacyl, and sinapyl monomers were reduced in almost the same ratio compared to the wild type. Our results suggest GH2 acts as a primarily multifunctional CAD to synthesize coniferyl and sinapyl alcohol precursors in rice lignin biosynthesis.

 首页
首页