

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (2): 298-307.DOI: 10.11983/CBB22240 cstr: 32102.14.CBB22240
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Rui Qiu1,2, Feng He2, Rui Li2, Yamei Wang2, Sinian Xing2, Yingping Cao2, Yefei Liu1,2, Xinyue Zhou1, Yan Zhao1,*( ), Chunxiang Fu2,*(
), Chunxiang Fu2,*( )
)
Received:2022-10-13
Accepted:2023-01-16
Online:2023-03-01
Published:2023-03-15
Contact:
*E-mail: Rui Qiu, Feng He, Rui Li, Yamei Wang, Sinian Xing, Yingping Cao, Yefei Liu, Xinyue Zhou, Yan Zhao, Chunxiang Fu. Highly Efficient Gene Editing of Lignin Gene F5H in Switchgrass[J]. Chinese Bulletin of Botany, 2023, 58(2): 298-307.
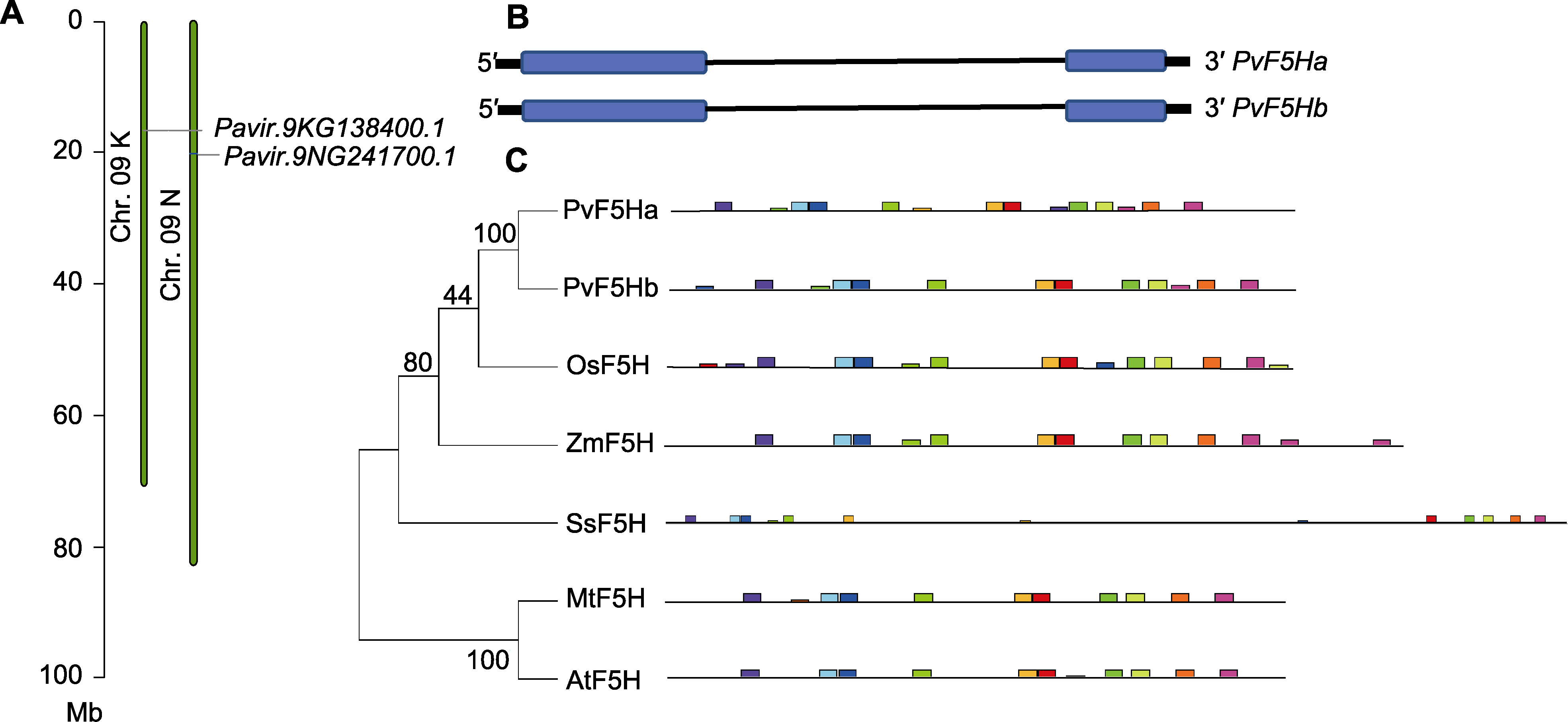
Figure 1 The basic molecular characterization of PvF5H in Panicum virgatum (A) Chromosomal position of F5H gene in P. virgatum (PvF5Ha: Pavir.9NG241700.1; PvF5Hb: Pavir.9KG138400.1); (B) F5H gene exons and introns distribution map of P. virgatum (blue boxes are exons, black thin lines are introns, thick black lines are 5' and 3' untranslated sequences); (C) Phylogenetic tree of F5H protein and gene conserved motif analysis in different species (P. virgatum: Pavir.9NG241700.1 (PvF5Ha), Pavir.9KG138400.1 (PvF5Hb); Oryza sativa: LOC_Os10g36848.1 (OsF5H); Zea mays: ZmPHB47.01G353500.1 (ZmF5H); Sorghum bicolor: Sobic.001G196300.1.p (SsF5H); Medicago truncatula: Medtr5g- 021390.1 (MtF5H); Arabidopsis thaliana: AT4G362201.1 (AtF5H). Different color of elements represent different nucleotide sequences in motif analysis)
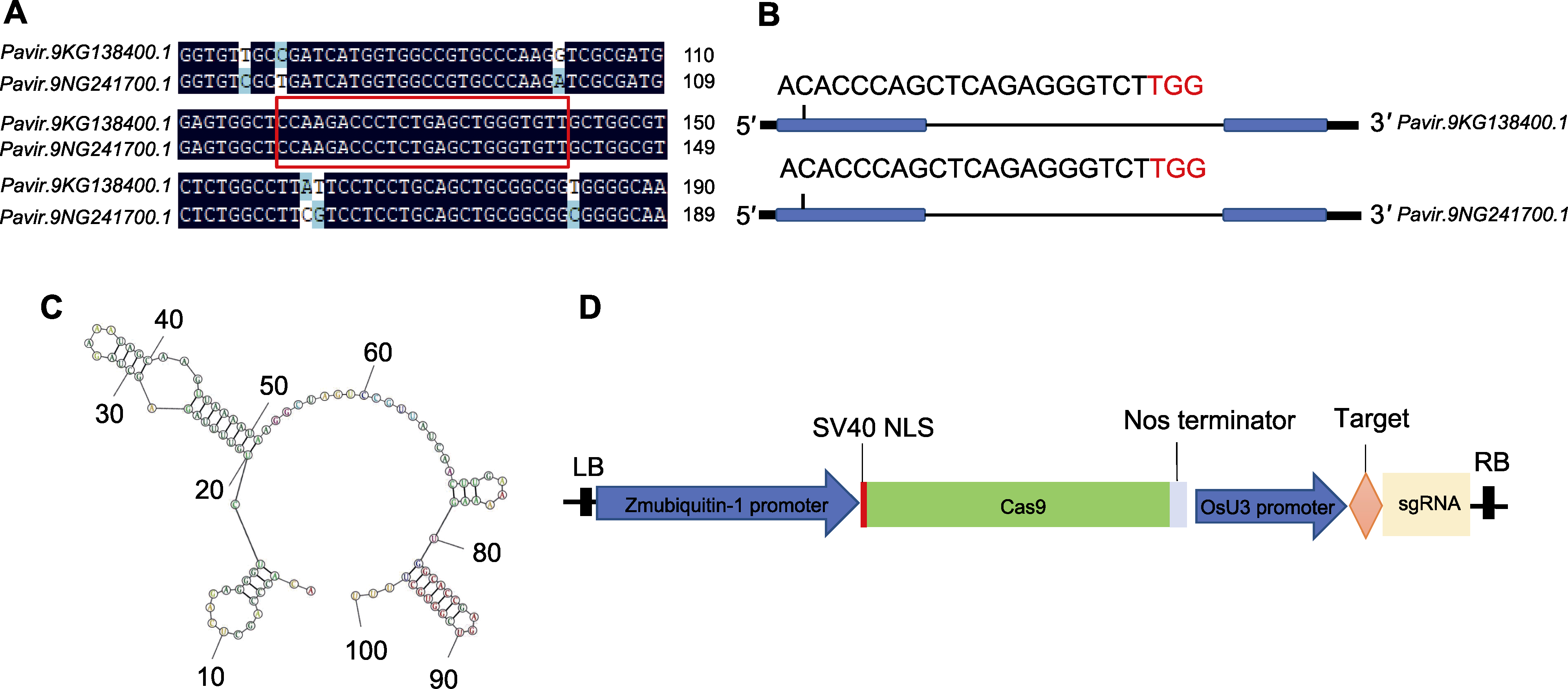
Figure 2 Construction of PvF5H gene editing vector in Panicum virgatum (A) PvF5H1a and PvF5H1b gene sequence alignment (red box indicates target site); (B) Schematic diagram of F5H gene editing site in switchgrass (blue for exons, thin lines for introns); (C) Secondary structure prediction map of F5H gRNA target+gRNA skeleton in switchgrass; (D) F5H gene editing vector map (the expression of Cas9 was driven by maize Ubiquitin promoter and sgRNA by OsU3 promoter)
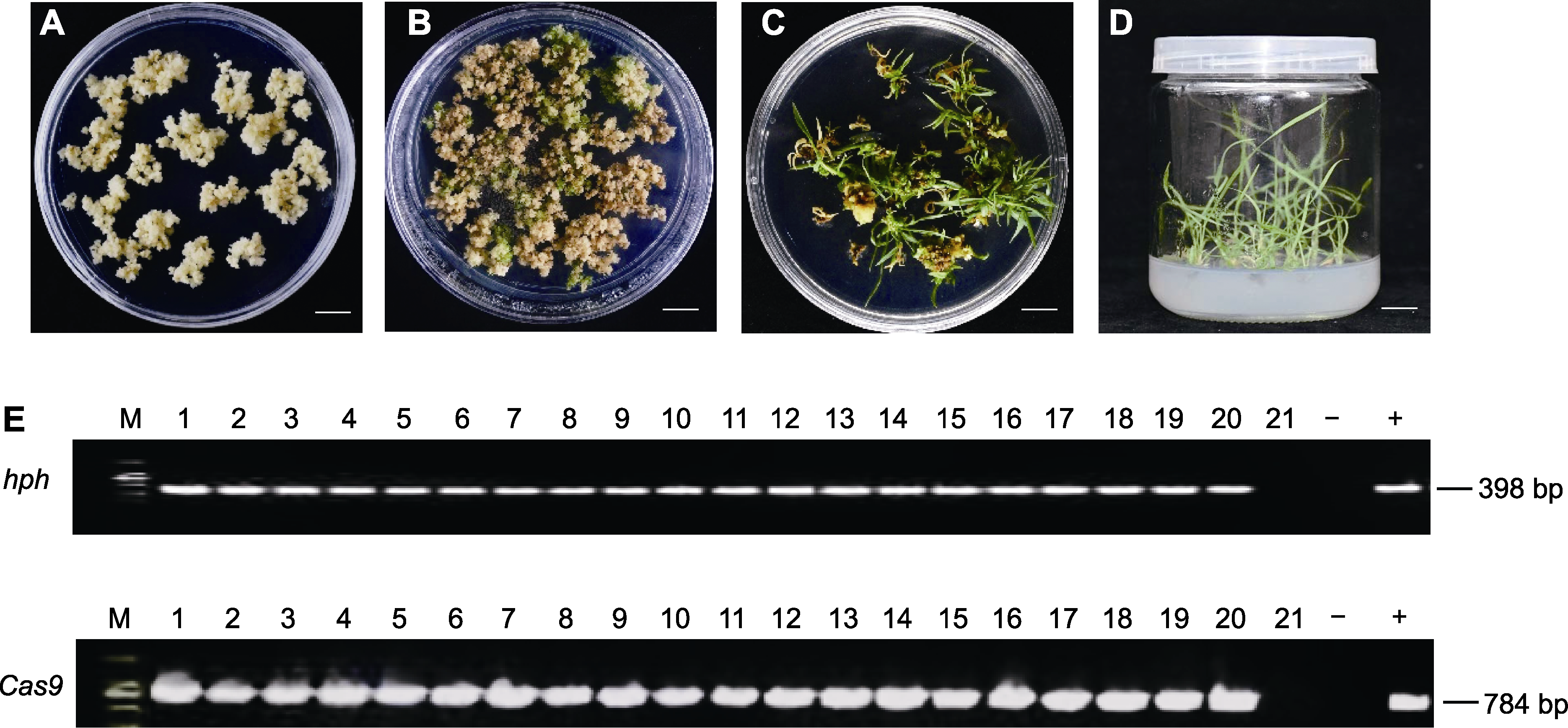
Figure 3 Generation and identification of CRISPR/Cas9_PvF5H transgenic switchgrass (A) Screening of switchgrass resistant calli grown on medium; (B) Calli turned green and produced shoots on differentiation medium after resistance screening; (C) Shoot enlongation; (D) Roots formation in rooting medium; (E) PCR analysis of transgenic switchgrass lines. No fragment band was amplified in the negative control (-, wild type switchgrass), while the same-sized band were amplified in all the resistant plants (lane 2 to 20) as in the positive control (+, CRISPR/Cas9_PvF5H plasmid). (A)-(D) Bars=1 cm
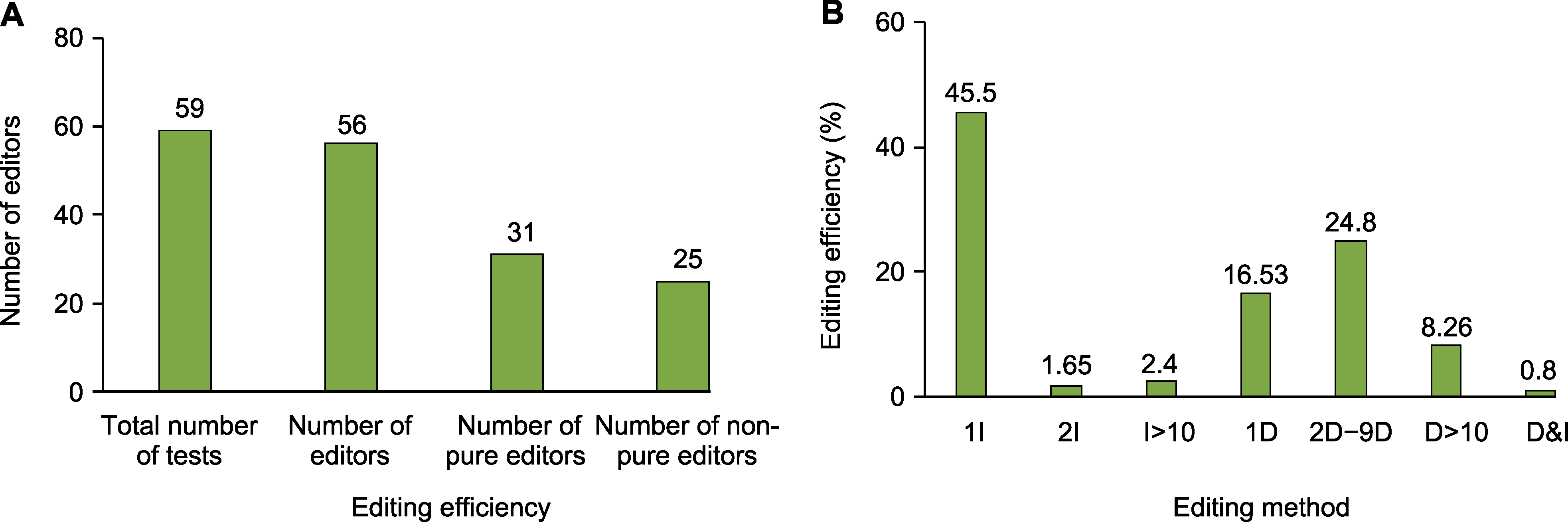
Figure 4 Gene editing methods and efficiency in switchgrass (A) Efficiency of F5H gene editing in switchgrass; (B) Gene editing patterns of PvF5H in switchgrass (I: Insertion; D: Delete; D&I: Delete+insertion)
| [1] |
何海锋, 吴娜, 刘吉利, 陈娟, 刘晓侠, 常雯雯 (2020). 柳枝稷种植年限对盐碱土壤理化性质的影响. 生态环境学报 29, 285-292.
DOI |
| [2] | 金枝, 陈倩, 代琳心, 马建锋 (2022). 木质纤维细胞壁大分子取向研究进展. 北京林业大学学报 44(12), 153-160. |
| [3] |
林萌萌, 李春娟, 闫彩霞, 孙全喜, 赵小波, 王娟, 苑翠玲, 单世华 (2021). CRISPR/Cas9基因编辑技术在作物中的应用. 核农学报 35, 1329-1339.
DOI |
| [4] | 刘吉利, 朱万斌, 谢光辉, 林长松, 程序 (2009). 能源作物柳枝稷研究进展. 草业学报 18(3), 232-240. |
| [5] | 马永清, 郝智强, 熊韶峻, 刘吉利 (2012). 我国柳枝稷规模化种植现状与前景. 中国农业大学学报 17(6), 133-137. |
| [6] | 王进 (2009). 亚麻(Linum usitatissimum)木质素合成关键酶基因的克隆及表达分析. 硕士论文. 北京: 中国农业科学院. pp. 3-5. |
| [7] | 徐超, 方志, 杨芳梅, 杨君祎, 闫冲冲, 邹鹤伟, 张金云, 林毅, 蔡永萍 (2015). 砀山酥梨果实F5H表达与石细胞发育的分析. 植物生理学报 51, 778-784. |
| [8] |
Akbas MY, Stark BC (2016). Recent trends in bioethanol production from food processing byproducts. J Ind Microbiol Biotechnol 43, 1593-1609.
DOI URL |
| [9] |
Amthor JS (2003). Efficiency of lignin biosynthesis: a quantitative analysis. Ann Bot 91, 673-695.
DOI URL |
| [10] |
Andersen JR, Zein I, Wenzel G, Darnhofer B, Eder J, Ouzunova M, Lübberstedt T (2008). Characterization of phenylpropanoid pathway genes within European maize (Zea mays L.) inbreds. BMC Plant Biol 8, 2.
DOI PMID |
| [11] |
Baskin TI (2017). Plant cell growth: cellulose caught slipping. Nat Plants 3, 17063.
DOI PMID |
| [12] | Baxter HL, Mazarei M, Labbe N, Kline LM, Cheng QK, Windham MT, Mann DGJ, Fu CX, Ziebell A, Sykes RW, Rodriguez M Jr, Davis MF, Mielenz JR, Dixon RA, Wang ZY, Stewart CN Jr (2014). Two-year field analysis of reduced recalcitrance transgenic switchgrass. Plant Bio- technol J 12, 914-924. |
| [13] |
Boerjan W, Ralph J, Baucher M (2003). Lignin biosynthesis. Annu Rev Plant Biol 54, 519-546.
PMID |
| [14] |
Bortesi L, Fischer R (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33, 41-52.
DOI PMID |
| [15] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020). TBtools: an integrative toolkit deveoped for interactive analyses of big biological data. Mol Plant 13, 1194-1202.
DOI URL |
| [16] |
Chen F, Dixon RA (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25, 759-761.
DOI PMID |
| [17] |
Fu CX, Mielenz JR, Xiao XR, Ge YX, Hamilton CY, Rodriguez M Jr, Chen F, Foston M, Ragauskas A, Bouton J, Dixon RA, Wang ZY (2011). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108, 3803-3808.
DOI PMID |
| [18] |
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13, 73-88.
DOI PMID |
| [19] |
Humphreys JM, Chapple C (2002). Rewriting the lignin roadmap. Curr Opin Plant Biol 5, 224-229.
DOI PMID |
| [20] | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Scien- ce 337, 816-821. |
| [21] |
Jung JH, Fouad WM, Vermerris W, Gallo M, Altpeter F (2012). RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol J 10, 1067-1076.
DOI PMID |
| [22] |
Keshwani DR, Cheng JJ (2009). Switchgrass for bioethanol and other value-added applications: a review. Bioresour Technol 100, 1515-1523.
DOI URL |
| [23] | Kim J, Choi B, Park YH, Cho BK, Lim HS, Natarajan S, Park SU, Bae H (2013). Molecular characterization of ferulate 5-hydroxylase gene from kenaf (Hibiscus cannabinus L.). Sci World J 2013, 421578. |
| [24] |
Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leplé JC, Boerjan W, Ferret V, De Nadai V, Jouanin L (1999). Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol 119, 153-164.
DOI PMID |
| [25] | Lovell JT, MacQueen AH, Mamidi S, Bonnette J, Jenkins J, Napier JD, Sreedasyam A, Healey A, Session A, Shu SQ, Barry K, Bonos S, Boston L, Daum C, Deshpande S, Ewing A, Grabowski PP, Haque T, Harrison M, Jiang JM, Kudrna D, Lipzen A, Pendergast IV TH, Plott C, Qi P, Saski CA, Shakirov EV, Sims D, Sharma M, Sharma R, Stewart A, Singan VR, Tang YH, Thibivillier S, Webber J, Weng XY, Williams M, Wu GA, Yoshinaga Y, Zane M, Zhang L, Zhang JY, Behrman KD, Boe AR, Fay PA, Fritschi FB, Jastrow JD, Lloyd-Reilley J, Martínez-Reyna JM, Matamala R, Mitchell RB, Rouquette FM Jr, Ronald P, Saha M, Tobias CM, Udvardi M, Wing RA, Wu YQ, Bartley LE, Casler M, Devos KM, Lowry DB, Rokhsar DS, Grimwood J, Juenger TE, Schmutz J (2021). Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature 590, 438-444. |
| [26] |
Meyer K, Cusumano JC, Somerville C, Chapple CC (1996). Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monoxygenases. Proc Natl Acad Sci USA 93, 6869-6874.
PMID |
| [27] | Nageswara-Rao M, Soneji JR, Kwit C, Stewart CN Jr (2013). Advances in biotechnology and genomics of swi- tchgrass. Biotechnol Biofuels 6, 77. |
| [28] |
Park JJ, Yoo CG, Flanagan A, Pu YQ, Debnath S, Ge YX, Ragauskas AJ, Wang ZY (2017). Defined tetra-allelic gene disruption of the 4-coumarate: coenzyme A ligase 1 (Pv4CL1) gene by CRISPR/Cas9 in switchgrass results in lignin reduction and improved sugar release. Biotechnol Biofuels 10, 284.
DOI |
| [29] |
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133, 1051-1071.
DOI URL |
| [30] |
Sakiroglu M, Sherman-Broyles S, Story A, Moore KJ, Doyle JJ, Brummer EC (2012). Patterns of linkage dise- quilibrium and association mapping in diploid alfalfa (M. sativa L.). Theor Appl Genet 125, 577-590.
DOI PMID |
| [31] |
Shen H, He XZ, Poovaiah CR, Wuddineh WA, Ma JY, Mann DGJ, Wang HZ, Jackson L, Tang YH, Neal Stewart C Jr, Chen F, Dixon RA (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193, 121-136.
DOI PMID |
| [32] | Shen H, Mazarei M, Hisano H, Escamilla-Trevino L, Fu CX, Pu YQ, Rudis MR, Tang YH, Xiao XR, Jackson L, Li GF, Hernandez T, Chen F, Ragauskas AJ, Stewart CN Jr, Wang ZY, Dixon RA (2013). A genomics approach to deciphering lignin biosynthesis in switchgrass. Plant Cell 25, 4342-4361. |
| [33] | Stewart JJ, Akiyama T, Chapple C, Ralph J, Mansfield SD (2009). The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Phy- siol 150, 621-635. |
| [34] | Wu ZY, Wang NF, Hisano H, Cao YP, Wu FY, Liu WW, Bao Y, Wang ZY, Fu CX (2019). Simultaneous regulation of F5H in COMT-RNAi transgenic switchgrass alterse- fects of COMT suppression on syringyl lignin biosynthe-sis. Plant Biotechnol J 17, 836-845. |
| [35] |
Xu B, Escamilla-Treviño LL, Sathitsuksanoh N, Shen ZX, Shen H, Zhang YHP, Dixon RA, Zhao BY (2011). Silencing of 4-coumarate: coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol 192, 611-625.
DOI URL |
| [1] | DU Shu-Hui, CHU Jian-Min, DUAN Jun-Guang, XUE Jian-Guo, XU Lei, XU Xiao-Qing, WANG Qi-Bing, HUANG Jian-Hui, ZHANG Qian. Influence of lignin phenols on soil organic carbon in degraded grassland in Nei Mongol, China [J]. Chin J Plant Ecol, 2025, 49(1): 30-41. |
| [2] | Jinyu Du, Zhen Sun, Yanlong Su, Heping Wang, Yaling Liu, Zhenying Wu, Feng He, Yan Zhao, Chunxiang Fu. Identification and Functional Analysis of an Agropyron mongolicum Caffeic Acid 3-O-methyltransferase Gene AmCOMT1 [J]. Chinese Bulletin of Botany, 2024, 59(3): 383-396. |
| [3] | Heping Wang, Zhen Sun, Yuchen Liu, Yanlong Su, Jinyu Du, Yan Zhao, Hongbo Zhao, Zhaoming Wang, Feng Yuan, Yaling Liu, Zhenying Wu, Feng He, Chunxiang Fu. Sequence Identification and Functional Analysis of Cinnamyl Alcohol Dehydrogenase Gene from Agropyron mongolicum [J]. Chinese Bulletin of Botany, 2024, 59(2): 204-216. |
| [4] | Wenqi Zhou, Yuqian Zhou, Yongsheng Li, Haijun He, Yanzhong Yang, Xiaojuan Wang, Xiaorong Lian, Zhongxiang Liu, Zhubing Hu. ZmICE2 Regulates Stomatal Development in Maize [J]. Chinese Bulletin of Botany, 2023, 58(6): 866-881. |
| [5] | Xian Deng, Tong Li, Xiaofeng Cao. Application and Prospect of Gene Editing in Forage Grass Breeding [J]. Chinese Bulletin of Botany, 2023, 58(2): 233-240. |
| [6] | He Xiaoling, Liu Pengcheng, Ma Bojun, Chen Xifeng. Advance in Gene-editing Technology Based on CRISPR/Cas9 and Its Application in Plants [J]. Chinese Bulletin of Botany, 2022, 57(4): 508-531. |
| [7] | Weitao Li, Min He, Xuewei Chen. Discovery of ZmFBL41 Chang7-2 as A Key Weapon against Banded Leaf and Sheath Blight Resistance in Maize [J]. Chinese Bulletin of Botany, 2019, 54(5): 547-549. |
| [8] | CHEN Hui-Ying, ZHANG Jing-Hui, HUANG Yong-Mei, GONG Ji-Rui. Traits related to carbon sequestration of common plant species in a Stipa grandis steppe in Nei Mongol under different land-uses [J]. Chin J Plant Ecol, 2014, 38(8): 821-832. |
| [9] | Lin Liu, Bin Yu, Pengyan Huang, Jun Jia, Hua Zhao, Junhua Peng, Peng Chen, Liangcai Peng. Frequency of Callus Induction and Plant Regeneration Among Eight Genotypes in Miscanthus sinensis Species [J]. Chinese Bulletin of Botany, 2013, 48(2): 192-198. |
| [10] | Shengchun Zhang, Changliang Ju, Xiaojing Wang. Arabidopsis Laccase Gene AtLAC4 Regulates Plant Growth and Responses to Abiotic Stress [J]. Chinese Bulletin of Botany, 2012, 47(4): 357-365. |
| [11] | Zhen Li;Hongzhi Wang;Ruifen Li;Jianhua Wei*. Lignin Biosynthesis and Manipulation in Plants and Utilization of Biomass Energy [J]. Chinese Bulletin of Botany, 2009, 44(03): 262-272. |
| [12] | Haiyong Liang Xiuying Xia Xiaorong Gao Qiao Su. Vector Construction with UGPase and anti4CL, and Their Expression in Transgenic Tobacco [J]. Chinese Bulletin of Botany, 2007, 24(04): 459-464. |
| [13] | HUANG Yao, SHEN Yu, ZHOU Mi, MA Rui-Sheng. Decomposition of Plant Residue as Influenced by its Lignin and N Itrogen [J]. Chin J Plan Ecolo, 2003, 27(2): 183-188. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||