

Chinese Bulletin of Botany ›› 2025, Vol. 60 ›› Issue (4): 515-532.DOI: 10.11983/CBB24112 cstr: 32102.14.CBB24112
• RESEARCH ARTICLES • Previous Articles Next Articles
Manya Zhao, Qiannan Sun, Jingjing Xu, Tianni Duan, Jintao Cai, Jing Zhou, Tingting Fan, Langtao Xiao*( ), Ruozhong Wang*(
), Ruozhong Wang*( )
)
Received:2024-07-23
Accepted:2025-06-04
Online:2025-07-10
Published:2025-06-04
Contact:
*E-mail: ltxiao@hunau.edu.cn;wangruozhong@hunau.edu.cn
Manya Zhao, Qiannan Sun, Jingjing Xu, Tianni Duan, Jintao Cai, Jing Zhou, Tingting Fan, Langtao Xiao, Ruozhong Wang. Identification, Mapping and Transcriptome Analysis of a New Leaf Color Mutant in Cucumber[J]. Chinese Bulletin of Botany, 2025, 60(4): 515-532.
| Primer name | Forward primer (5′→3′) | Reverse primer (5'→3') |
|---|---|---|
| Cs-Actin | GTTACGCCCTCCCTCATGCCATTC | TCCCGTTCGGCAGTGGTGGT |
| CsaV3_3G002180 | GTCGTCCTGCCATTCGATCA | AGCACCAAGTTCACTCCAACT |
| CsaV3_5G025230 | AATTCTTCCGACCCGAACCC | AGTAGCCTTCTGCGGACCTA |
| CsaV3_1G030370 | CAGTGGCTGGATACGTCCTC | GTGAGCTCCCGCCATAAAGT |
| CsaV3_7G000620 | TGGAGCATCTCCGAAAGTGG | GGCAAGGAATTGTGATGCCA |
| CsaV3_5G028880 | TAGAACCCAGGCTCCCTCAA | CCGTGTTTTCACAAGCTTCTCT |
| CsaV3_3G041340 | AACATGTTACTGGTGGGGGC | CACATTGAAATCATTGGGTACCTG |
| CsaV3_6G037230 | CCCACTCAAGCGATGTG CTA | CCATTGACCTCAGCATTGCG |
| CsaV3_5G006200 | GACCCAGTTCAAGCTAGCCA | ACTGAGACAACAAGCGCGTA |
| CsaV3_7G008610 | GGCTCCAAGGGCCAATACAT | GTAGGCCTTGGACAGGCATT |
Table 1 Primers used for qRT-PCR
| Primer name | Forward primer (5′→3′) | Reverse primer (5'→3') |
|---|---|---|
| Cs-Actin | GTTACGCCCTCCCTCATGCCATTC | TCCCGTTCGGCAGTGGTGGT |
| CsaV3_3G002180 | GTCGTCCTGCCATTCGATCA | AGCACCAAGTTCACTCCAACT |
| CsaV3_5G025230 | AATTCTTCCGACCCGAACCC | AGTAGCCTTCTGCGGACCTA |
| CsaV3_1G030370 | CAGTGGCTGGATACGTCCTC | GTGAGCTCCCGCCATAAAGT |
| CsaV3_7G000620 | TGGAGCATCTCCGAAAGTGG | GGCAAGGAATTGTGATGCCA |
| CsaV3_5G028880 | TAGAACCCAGGCTCCCTCAA | CCGTGTTTTCACAAGCTTCTCT |
| CsaV3_3G041340 | AACATGTTACTGGTGGGGGC | CACATTGAAATCATTGGGTACCTG |
| CsaV3_6G037230 | CCCACTCAAGCGATGTG CTA | CCATTGACCTCAGCATTGCG |
| CsaV3_5G006200 | GACCCAGTTCAAGCTAGCCA | ACTGAGACAACAAGCGCGTA |
| CsaV3_7G008610 | GGCTCCAAGGGCCAATACAT | GTAGGCCTTGGACAGGCATT |
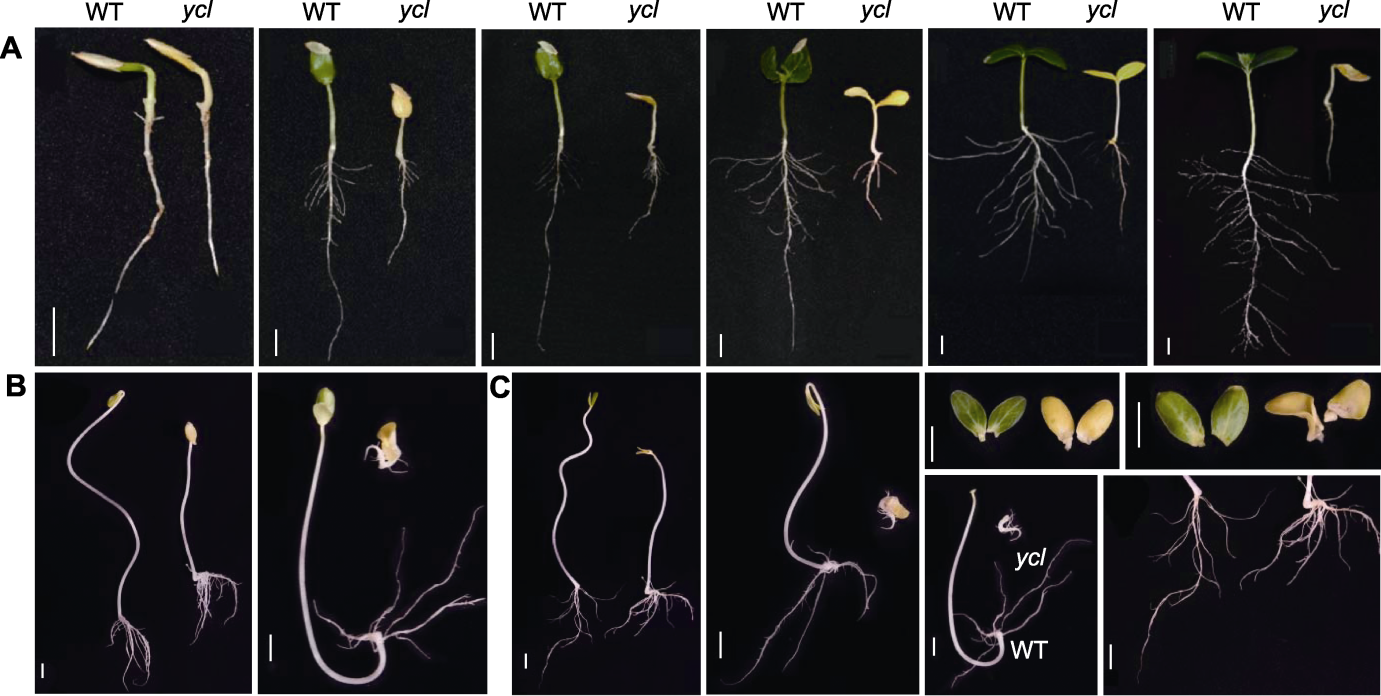
Figure 1 Comparison of phenotypic changes between ycl and XYYH-3-1 at the cotyledon stage of cucumber (A) Phenotypic changes of wild type (WT) and ycl in 4, 5, 6, 7, 8 and 14 days (natural light); (B) Phenotypic differences of WT and ycl between the two groups at 7 d of age (dark treatment for 7 d); (C) Phenotypic differences of WT and ycl between the two groups at 8 d of age (light culture for 1 d). Bars=1 cm
| Material | Plant height (cm) | Root length (cm) | Stem diameter (cm) | Leaf area (cm2) |
|---|---|---|---|---|
| WT | 4.86±0.32 | 15.56±0.94 | 0.22±0.03 | 9.04±1.66 |
| ycl | 2.64±0.56** | 5.5±0.9** | 0.18±0.02* | 1.95±0.26** |
Table 2 Agronomic characters of ycl and XYYH-3-1 at cotyledon stage (7 d) of cucumber
| Material | Plant height (cm) | Root length (cm) | Stem diameter (cm) | Leaf area (cm2) |
|---|---|---|---|---|
| WT | 4.86±0.32 | 15.56±0.94 | 0.22±0.03 | 9.04±1.66 |
| ycl | 2.64±0.56** | 5.5±0.9** | 0.18±0.02* | 1.95±0.26** |
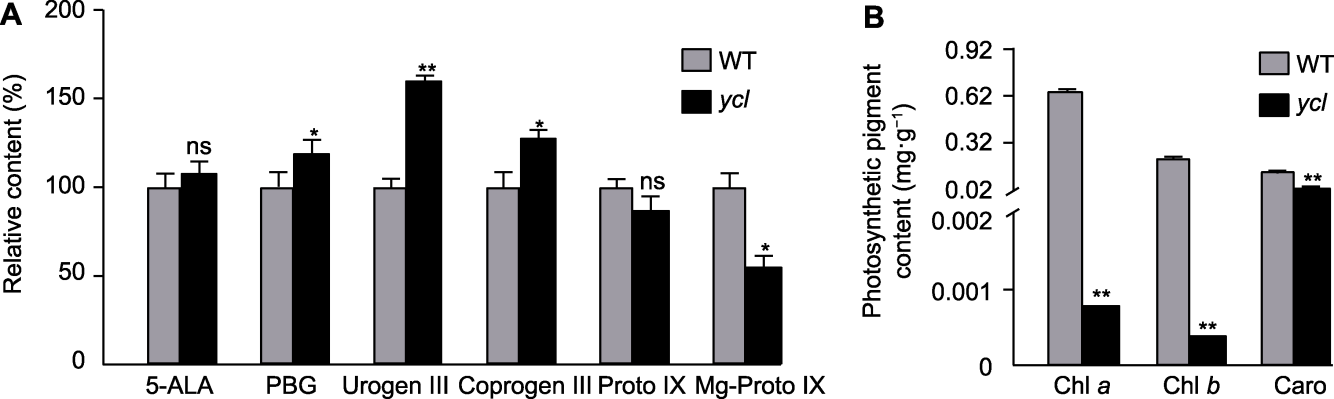
Figure 2 Content of chlorophyll and its biosynthetic intermediate metabolites in ycl and XYYH-3-1 (A) Relative content of chlorophyll biosynthetic intermediate metabolites; (B) Photosynthetic pigment content. WT: Wild type; 5-ALA: 5-aminolevulinic acid; PBG: Porphobilinogen; Urogen III: Uroporphyrinogen III; Coprogen III: Coproporphyrinogen III; Proto IX: Protoporphyrin IX; Mg-Proto IX: Mg-protoporphyrin IX. * denote significant differences (t-test, P<0.05); ** denote extremely significant differences (t-test, P<0.01); ns denote no significant difference.
| Material | Chl a (mg·g−1) | Chl b (mg·g−1) | Caro (mg·g−1) | Total Chl (mg·g−1) | Chl a/b | Total Chl/Caro |
|---|---|---|---|---|---|---|
| WT | 0.6483±0.0162 | 0.2157±0.0063 | 0.1304±0.0018 | 0.8641±0.0225 | 3.0057 | 6.6272 |
| ycl | 0.0008±0** | 0.0004±0** | 0.0265±0.0007** | 0.0012±0** | 1.9303 | 0.0443 |
Table 3 Photosynthetic pigment content in leaves of ycl and XYYH-3-1
| Material | Chl a (mg·g−1) | Chl b (mg·g−1) | Caro (mg·g−1) | Total Chl (mg·g−1) | Chl a/b | Total Chl/Caro |
|---|---|---|---|---|---|---|
| WT | 0.6483±0.0162 | 0.2157±0.0063 | 0.1304±0.0018 | 0.8641±0.0225 | 3.0057 | 6.6272 |
| ycl | 0.0008±0** | 0.0004±0** | 0.0265±0.0007** | 0.0012±0** | 1.9303 | 0.0443 |
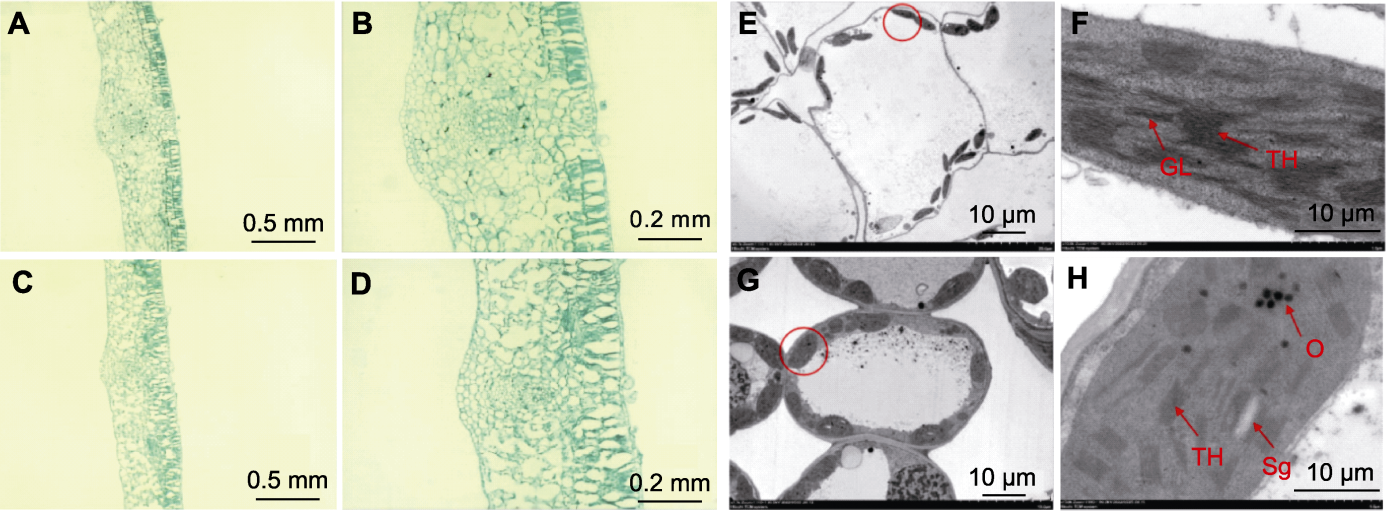
Figure 3 Microstructure and chloroplast ultrastructure of ycl and XYYH-3-1 leaves (A), (B) Wild type cotyledon microstructures; (C), (D) Mutant cotyledon microstructures; (E), (F) Wild type chloroplast ultrastructures; (G), (H) ycl chloroplast ultrastructures. (F) is an enlarged version of the chloroplast in the red circle in (E). (H) is an enlarged version of the chloroplast in the red circle in (G). Sg: Starch grain; TH: Thylakoid; GL: Grana lamella; O: Osmiophilic body
| Material | Pn (μmol·m-2·s-1) | Gs (mmol·m-2·s-1) | Ci (μmol·mol-1) | Tr (mol·m-2·s-1) |
|---|---|---|---|---|
| WT | 15.85±1.39 | 0.37±0.08 | 331±12 | 4.53±0.62 |
| ycl | -2.49±0.15** | 0.14±0.02** | 458±7** | 2.17±0.31** |
Table 4 Photosynthesis parameters of the ycl and XYYH-3-1
| Material | Pn (μmol·m-2·s-1) | Gs (mmol·m-2·s-1) | Ci (μmol·mol-1) | Tr (mol·m-2·s-1) |
|---|---|---|---|---|
| WT | 15.85±1.39 | 0.37±0.08 | 331±12 | 4.53±0.62 |
| ycl | -2.49±0.15** | 0.14±0.02** | 458±7** | 2.17±0.31** |
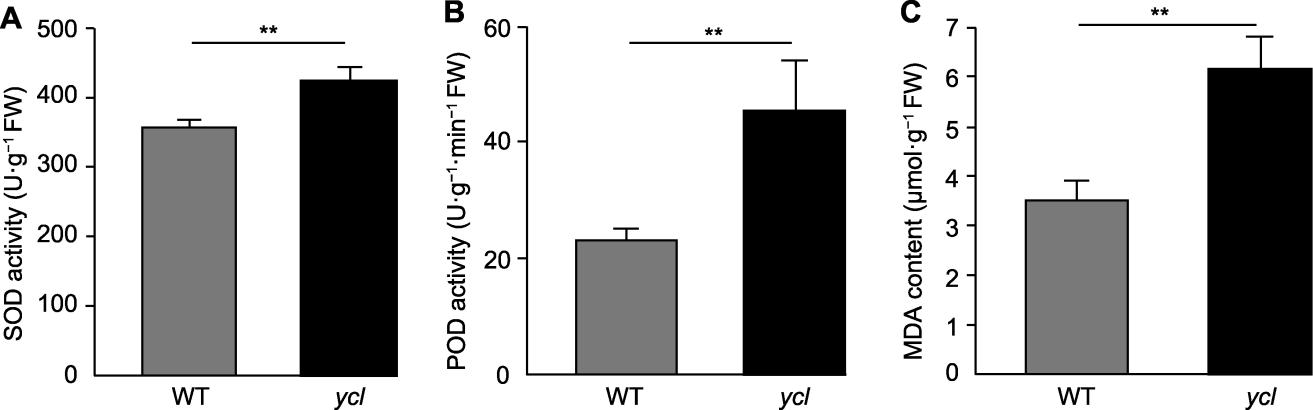
Figure 4 Antioxidant enzyme activities and malondialdehyde contents of ycl and XYYH-3-1 (A) Superoxide dismutase (SOD) activity; (B) Peroxidase (POD) activity; (C) Malondialdehyde (MDA) contents. WT: Wild type. ** denote extremely significant differences (t-test, P<0.01).
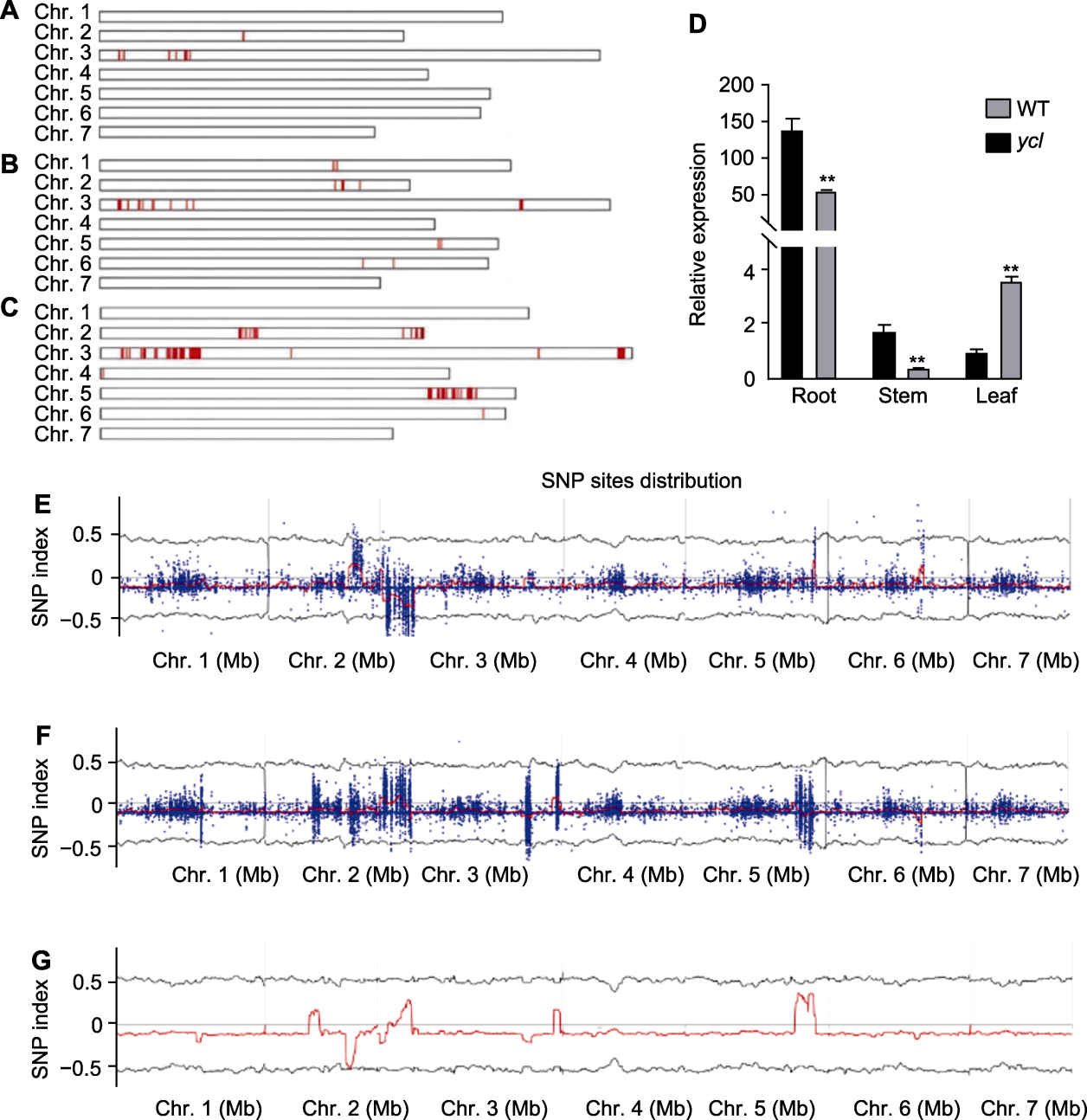
Figure 5 Preliminary localisation and screening of mutant genes (A)-(C) Distribution of single nucleotide polymorphisms ((A) Population 18; (B) XY cotyledon population; (C) XY true leaf population); (D) Relative expression of CsaV3_3G001980 (** denote extremely significant differences, t-test, P<0.01); (E)-(G) SNP index plot ((E) Population 18; (F) XY cotyledon population; (G) XY true leaf population). WT: Wild type; SNP: Single nucleotide polymorphism
| Genes within the interval | Annotation |
|---|---|
| CsaV3_3G001960 | Cytochrome P450 family ent-kaurenoic acid oxidase |
| CsaV3_3G001970 | Cytochrome P450 family ent-kaurenoic acid oxidase |
| CsaV3_3G001980 | Protein NRT1/PTR FAMILY 7.3-like |
| CsaV3_3G001990 | Hsp90 co-chaperone Cdc37, N-terminally processed like |
| CsaV3_3G002000 | Ribose-phosphate pyrophosphokinase |
| CsaV3_3G002010 | Type I inositol polyphosphate 5-phosphatase, puta |
| CsaV3_3G002020 | Phenylalanine-tRNA ligase alpha subunit like |
| CsaV3_3G002030 | Two-component response regulator-like aprr2 |
| CsaV3_3G002040 | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| CsaV3_3G002050 | Vacuolar protein sorting-associated protein 55 homolog |
| CsaV3_3G002060 | Haloacid dehalogenase-like hydrolase |
| CsaV3_3G002070 | Haloacid dehalogenase-like hydrolase domain-containing protein |
| CsaV3_3G002080 | Regulator of nonsense transcripts 1 homolog |
| CsaV3_3G002090 | Lon protease homolog 2, peroxisomal |
| CsaV3_3G002100 | Electron carrier/iron ion-binding protein |
| CsaV3_3G002110 | BOI-related E3 ubiquitin-protein ligase 1 |
| CsaV3_3G002120 | Protein of unknown function (DUF581) |
| CsaV3_3G002130 | Poly(U)-specific endoribonuclease |
| CsaV3_3G002140 | Methyltransferase-like protein 13 |
| CsaV3_3G002150 | Beta-amylase |
| CsaV3_3G002160 | Protein kinase, putative |
| CsaV3_3G002170 | Novel plant snare, putative |
| CsaV3_3G002180 | Phosphatidylinositol 4-phosphate 5-kinase 1 |
| CsaV3_3G002190 | Coiled-coil domain-containing protein 97 |
| CsaV3_3G002200 | Calcineurin B-like protein |
| CsaV3_3G002210 | Unknown protein |
| CsaV3_3G002220 | Protein kinase |
| CsaV3_3G002230 | Unknown protein |
| CsaV3_3G002240 | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain-containing protein |
| CsaV3_3G002250 | Peptidyl-prolyl cis-trans isomerase-like |
| CsaV3_3G002260 | DVA-1 polyprotein |
| CsaV3_3G002270 | tRNA/rRNA methyltransferase (SpoU) family protein |
| CsaV3_3G002280 | Protein lingerer like |
| CsaV3_3G002290 | Chlorophyll a/b binding family protein |
| CsaV3_3G002300 | ATP-dependent Clp protease proteolytic subunit |
| CsaV3_3G002310 | Unknown protein |
| CsaV3_3G002320 | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6-like |
| CsaV3_3G002330 | Kelch repeat-containing protein |
| CsaV3_3G002340 | GATA transcription factor 26-like |
| CsaV3_3G002350 | Vesicle-associated protein 2-1 |
| CsaV3_3G002360 | 50S ribosomal protein L19, chloroplastic |
Table 5 The genes within the preliminarily mapped region of ycl mutant site
| Genes within the interval | Annotation |
|---|---|
| CsaV3_3G001960 | Cytochrome P450 family ent-kaurenoic acid oxidase |
| CsaV3_3G001970 | Cytochrome P450 family ent-kaurenoic acid oxidase |
| CsaV3_3G001980 | Protein NRT1/PTR FAMILY 7.3-like |
| CsaV3_3G001990 | Hsp90 co-chaperone Cdc37, N-terminally processed like |
| CsaV3_3G002000 | Ribose-phosphate pyrophosphokinase |
| CsaV3_3G002010 | Type I inositol polyphosphate 5-phosphatase, puta |
| CsaV3_3G002020 | Phenylalanine-tRNA ligase alpha subunit like |
| CsaV3_3G002030 | Two-component response regulator-like aprr2 |
| CsaV3_3G002040 | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| CsaV3_3G002050 | Vacuolar protein sorting-associated protein 55 homolog |
| CsaV3_3G002060 | Haloacid dehalogenase-like hydrolase |
| CsaV3_3G002070 | Haloacid dehalogenase-like hydrolase domain-containing protein |
| CsaV3_3G002080 | Regulator of nonsense transcripts 1 homolog |
| CsaV3_3G002090 | Lon protease homolog 2, peroxisomal |
| CsaV3_3G002100 | Electron carrier/iron ion-binding protein |
| CsaV3_3G002110 | BOI-related E3 ubiquitin-protein ligase 1 |
| CsaV3_3G002120 | Protein of unknown function (DUF581) |
| CsaV3_3G002130 | Poly(U)-specific endoribonuclease |
| CsaV3_3G002140 | Methyltransferase-like protein 13 |
| CsaV3_3G002150 | Beta-amylase |
| CsaV3_3G002160 | Protein kinase, putative |
| CsaV3_3G002170 | Novel plant snare, putative |
| CsaV3_3G002180 | Phosphatidylinositol 4-phosphate 5-kinase 1 |
| CsaV3_3G002190 | Coiled-coil domain-containing protein 97 |
| CsaV3_3G002200 | Calcineurin B-like protein |
| CsaV3_3G002210 | Unknown protein |
| CsaV3_3G002220 | Protein kinase |
| CsaV3_3G002230 | Unknown protein |
| CsaV3_3G002240 | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain-containing protein |
| CsaV3_3G002250 | Peptidyl-prolyl cis-trans isomerase-like |
| CsaV3_3G002260 | DVA-1 polyprotein |
| CsaV3_3G002270 | tRNA/rRNA methyltransferase (SpoU) family protein |
| CsaV3_3G002280 | Protein lingerer like |
| CsaV3_3G002290 | Chlorophyll a/b binding family protein |
| CsaV3_3G002300 | ATP-dependent Clp protease proteolytic subunit |
| CsaV3_3G002310 | Unknown protein |
| CsaV3_3G002320 | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6-like |
| CsaV3_3G002330 | Kelch repeat-containing protein |
| CsaV3_3G002340 | GATA transcription factor 26-like |
| CsaV3_3G002350 | Vesicle-associated protein 2-1 |
| CsaV3_3G002360 | 50S ribosomal protein L19, chloroplastic |
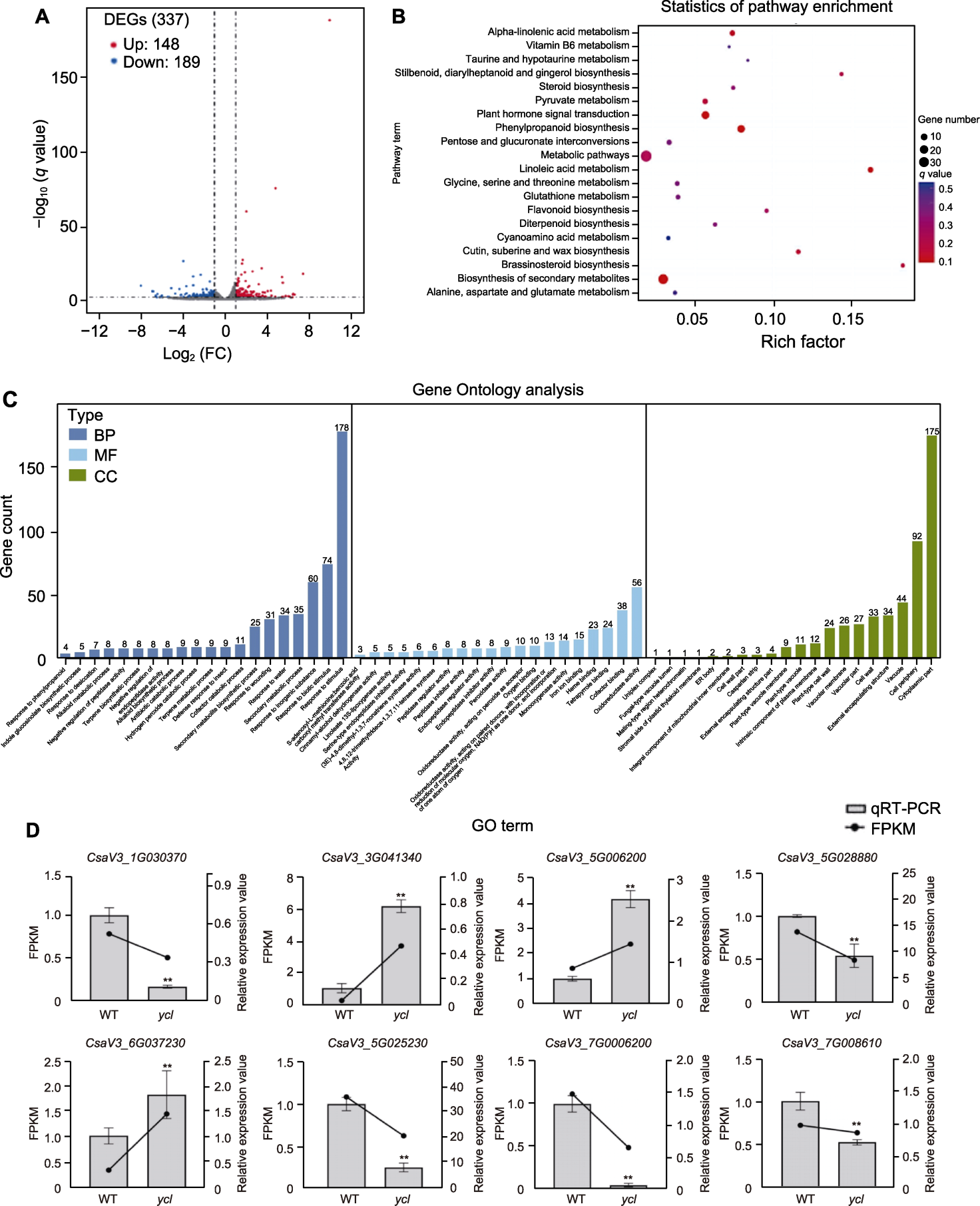
Figure 6 RNA-Seq analysis of differential expression between ycl and XYYH-3-1 (A) Differentially expressed genes volcano map; (B) Scatter plot of differentially expressed genes KEGG pathway enrichment; (C) GO term secondary classification statistics; (D) The qRT-PCR verification of transcriptome results. BP: Biological process; MF: Molecular function; CC: Cellular component; WT: Wild type; DEG: Differential expressed gene; FPKM: Fragments per kilobase per million mapped reads; FC: Fold Change. ** denote extremely significant differences (t-test, P<0.01).
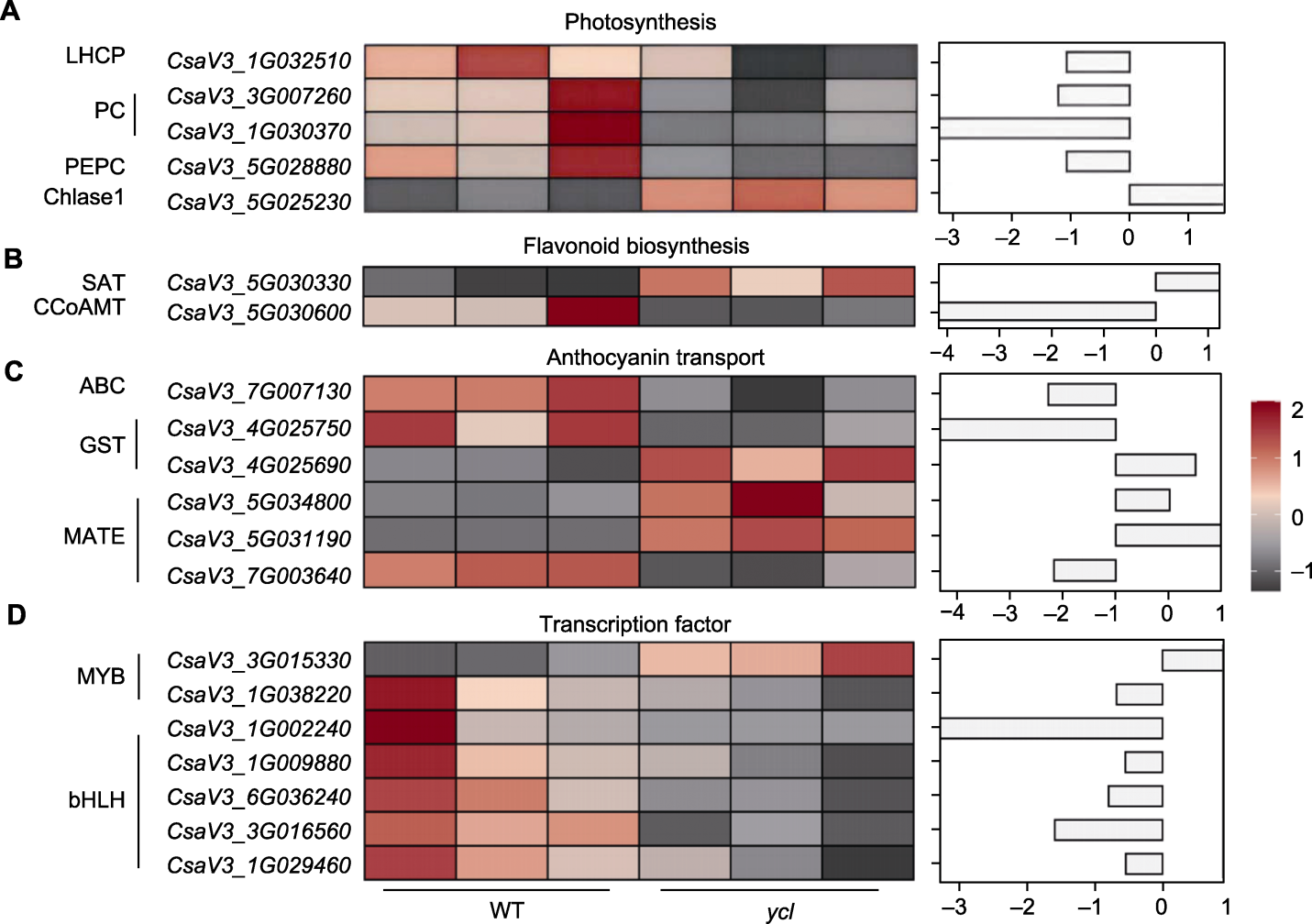
Figure 7 Differentially expressed genes (DEGs) associated with the ycl phenotype (A) DEGs involved in photosynthesis; (B) DEGs involved in flavonoid biosynthesis; (C) DEGs involved in anthocyanin transport; (D) DEGs encoding transcription factors. WT: Wild type; LHCP: Chlorophyll a/b binding protein; PC: Plastocyanin; PEPC: Phosphoenolpyruvate carboxylase; Chlase1: Chlorophyllase 1; SAT: Stemmadenine O-acetyltransferase; CCoAMT: Trans- caffeoyl-CoA 3-O-methyltransferase; GST: Glutathione-S-transferase; MATE: Multidrug and toxic compound extrusion
| [1] |
Allan AC, Hellens RP, Laing WA (2008). MYB transcription factors that colour our fruit. Trends Plant Sci 13, 99-102.
DOI PMID |
| [2] | Amaresh, Krishnan SG, Vinod KK, Chinnusamy V, Sevanthi ACRMV, Dhandapani R, Ellur RK, Bhowmick PK, Bollinedi H, Umadevi P, Senapati M, Verma RK, Nagarajan M, Dhawan G, Kumar P, Singh AK (2023). Phenotypic characterization of the novel seedling stage zebra leaf mutant, Pusa Zebra 18 in rice. Plant Physiol Rep 28, 500-512. |
| [3] |
Awai K, Xu CC, Tamot B, Benning C (2006). A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc Natl Acad Sci USA 103, 10817-10822.
DOI PMID |
| [4] | Boase MR, Brendolise C, Wang L, Ngo H, Espley RV, Hellens RP, Schwinn KE, Davies KM, Albert NW (2015). Failure to launch: the self-regulating Md-MYB10R6 gene from apple is active in flowers but not leaves of Petunia. Plant Cell Rep 34, 1817-1823. |
| [5] | Bogorad L (1962). Porphyrin synthesis. Methods Enzymol 5, 885-895. |
| [6] |
Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, Martin C (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26, 1301-1308.
DOI PMID |
| [7] | Chen H, Cheng ZJ, Ma XD, Wu H, Liu YL, Zhou KN, Chen YL, Ma WW, Bi JC, Zhang X, Guo XP, Wang JL, Lei CL, Wu FQ, Lin QB, Liu YQ, Liu LL, Jiang L (2013). A knockdown mutation of YELLOW-GREEN LEAF2blocks chlorophyll biosynthesis in rice. Plant Cell Rep 32, 1855-1867. |
| [8] | Chen JY, Zhao J, Liu X, Li C, Lin DZ, Dong YJ, Ye SH, Zhang XM (2010). Genetic analysis and molecular mapping of a new thermosensitive leaf-color mutant in Oryza sativa. Chin Bull Bot 45, 419-425. (in Chinese) |
|
陈佳颖, 赵剑, 刘晓, 李超, 林冬枝, 董彦君, 叶胜海, 张小明 (2010). 一个新水稻温敏感叶色突变体的遗传分析及其基因分子定位. 植物学报 45, 419-425.
DOI |
|
| [9] | Cheng MZ, Meng FY, Mo FL, Chen XL, Zhang H, Wang AX (2022). Insights into the molecular basis of a yellow leaf color mutant (ym) in tomato (Solanum lycopersicum). Sci Hortic 293, 110743. |
| [10] |
Dasgupta K, Thilmony R, Stover E, Oliveira ML, Thomson J (2017). Novel R2R3-MYB transcription factors from Prunus americana regulate differential patterns of anthocyanin accumulation in tobacco and citrus. GM Crops Food 8, 85-105.
DOI PMID |
| [11] | Dei M (1985). Benzyladenine-induced stimulation of 5-aminolevulinic acid accumulation under various light intensities in levulinic acid-treated cotyledons of etiolated cucumber. Physiol Plant 64, 153-160. |
| [12] | Deng LC, Qin P, Liu Z, Wang GL, Chen WL, Tong JH, Xiao LT, Tu B, Sun YT, Yan W, He H, Tan J, Chen XW, Wang YP, Li SG, Ma BT (2017). Characterization and fine-mapping of a novel premature leaf senescence mutant yellow leaf and dwarf 1in rice. Plant Physiol Biochem 111, 50-58. |
| [13] | Ding Y, Yang W, Su CG, Ma HH, Pan Y, Zhang XG, Li JH (2019). Tandem 13-lipoxygenase genes in a cluster confers yellow-green leaf in cucumber. Int J Mol Sci 20, 3102. |
| [14] | Du WK, Hu FR, Yuan SX, Liu C (2020). The identification of key candidate genes mediating yellow seedling lethality in a Lilium regale mutant. Mol Biol Rep 47, 2487-2499. |
| [15] | Gao ML, Hu LL, Li YH, Weng YQ (2016). The chlorophyll-deficient golden leaf mutation in cucumber is due to a single nucleotide substitution in CsChlI for magnesium chelatase I subunit. Theor Appl Genet 129, 1961-1973. |
| [16] | Gao YF, Zhao DH, Zhang JQ, Chen JS, Li JL, Weng Z, Rong LP (2021). De novo transcriptome sequencing and anthocyanin metabolite analysis reveals leaf color of Acer pseudosieboldianum in autumn. BMC Genomics 22, 383. |
| [17] |
Ghangal R, Rajkumar MS, Garg R, Jain M (2020). Genome-wide analysis of glutathione S-transferase gene family in chickpea suggests its role during seed development and abiotic stress. Mol Biol Rep 47, 2749-2761.
DOI PMID |
| [18] | Guan HY, Xu XB, He CM, Liu CX, Liu Q, Dong R, Liu TS, Wang LM (2016). Fine mapping and candidate gene analysis of the leaf-color gene ygl-1 in maize. PLoS One 11, e0153962. |
| [19] | Han HW, Zhou Y, Liu HF, Chen XJ, Wang Q, Zhuang HM, Sun XX, Ling QH, Zhang HJ, Wang BK, Wang J, Tang YP, Wang H, Liu HY (2023). Transcriptomics and metabolomics analysis provides insight into leaf color and photosynthesis variation of the yellow-green leaf mutant of hami melon (Cucumis melo L.). Plants 12, 1623. |
| [20] | Hu LL, Zhang HQ, Xie C, Wang J, Zhang JY, Wang H, Weng YQ, Chen P, Li YH (2020). A mutation in CsHD encoding a histidine and aspartic acid domain-containing protein leads to yellow young leaf-1 (yyl-1) in cucumber (Cucumis sativus L.). Plant Sci 293, 110407. |
| [21] | Huang SN, Liu ZY, Li DY, Yao RP, Hou L, Li X, Feng H (2016). Physiological characterization and comparative transcriptome analysis of a slow-growing reduced-thylakoid mutant of Chinese cabbage (Brassica campestris ssp. pekinensis). Front Plant Sci 7, 3. |
| [22] | Huang WF, Zhang Y, Shen LQ, Fang Q, Liu Q, Gong CB, Zhang C, Zhou Y, Mao C, Zhu YL, Zhang JH, Chen HP, Zhang Y, Lin YJ, Bock R, Zhou F (2020). Accumulation of the RNA polymerase subunit RpoB depends on RNA editing by OsPPR16 and affects chloroplast development during early leaf development in rice. New Phytol 228, 1401-1416. |
| [23] | Jiang SH, Chen M, He NB, Chen XL, Wang N, Sun QG, Zhang TL, Xu HF, Fang HC, Wang YC, Zhang ZY, Wu SJ, Chen XS (2019). MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in ap- ple. Hortic Res 6, 40. |
| [24] |
Jiao FC, Zhao L, Wu XF, Song ZB, Li YP (2020). Metabolome and transcriptome analyses of the molecular mechanisms of flower color mutation in tobacco. BMC Genomics 21, 611.
DOI PMID |
| [25] | Ladygin VG (2006). Spectral features and structure of chloroplasts under an early block of chlorophyll synthesis. Bio- physics 51, 635-644. |
| [26] | Li CH, Zhu LH, Yang HJ, Song RH, Gu WH (2019). Main agronomic characters and biochemical traits of xantha mutant of vegetable soybean. Mol Plant Breed 17, 3726-3734. (in Chinese) |
| 李超汉, 朱丽华, 杨红娟, 宋荣浩, 顾卫红 (2019). 菜用大豆黄化新突变体的主要农艺性状和生理特性. 分子植物育种 17, 3726-3734. | |
| [27] | Li XF, Zhang ZL (2016). Plant Physiology Laboratory Manual (5th edn). Beijing: Science Press. pp. 30-31. (in Chinese) |
| 李小方, 张志良 (2016). 植物生理学实验指导(第5版). 北京: 科学出版社. pp. 30-31. | |
| [28] |
Lin N, Gao YM, Zhou QY, Ping XK, Li JN, Liu LZ, Yin JM (2022). Genetic mapping and physiological analysis of chlorophyll-deficient mutant in Brassica napus L. BMC Plant Biol 22, 244.
DOI PMID |
| [29] | Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, Gojon A, Tsay YF (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514-2528. |
| [30] | Liu J, Wang JY, Yao XY, Zhang Y, Li JQ, Wang XX, Xu ZJ, Chen WF (2015). Characterization and fine mapping of thermo-sensitive chlorophyll deficit mutant1 in rice (Oryza sativa L.). Breed Sci 65, 161-169. |
| [31] | Liu L, Sun TT, Liu XY, Guo Y, Huang X, Gao P, Wang XZ (2019). Genetic analysis and mapping of a striped rind gene (st3) in melon (Cucumis melo L.). Euphytica 215, 20. |
| [32] | Liu P, Li MJ (2016). Experiments in Plant Physiology (2nd edn). Beijing: Science Press. pp. 142-150. (in Chinese) |
| 刘萍, 李明军 (2016). 植物生理学实验(第2版). 北京: 科学出版社. pp. 142-150. | |
| [33] | Liu X, Huang QQ, Yang YR, Tang JY, Zhao YN, Zhang J (2021). Characterization and map-based cloning of the novel rice yellow leaf mutant yl3. J Plant Biol 64, 35-44. |
| [34] | Mao GZ, Wei HL, Hu W, Ma Q, Zhang M, Wang HT, Yu SX (2019). Fine mapping and molecular characterization of the virescent gene vsp in upland cotton (Gossypium hirsutum). Theor Appl Genet 132, 2069-2086. |
| [35] | Miao H, Zhang SP, Wang M, Wang Y, Weng YQ, Gu XF (2016). Fine mapping of virescent leaf gene v-1 in cucumber (Cucumis sativus L.). Int J Mol Sci 17, 1602. |
| [36] |
Michelmore RW, Paran I, Kesseli RV (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88, 9828-9832.
DOI PMID |
| [37] | Nagata N, Tanaka R, Satoh S, Tanaka A (2005). Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of prochlorococcus species. Plant Cell 17, 233-240. |
| [38] | Parks BM, Quail PH (1991). Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3, 1177-1186. |
| [39] | Pierce LK, Wehner TC (1990). Review of genes and linkage groups in cucumber. Hortscience 25, 605-615. |
| [40] | Qin R, Zeng DD, Liang R, Yang CC, Akhter D, Alamin M, Jin XL, Shi CH (2017). Rice gene SDL/RNRS1, encoding the small subunit of ribonucleotide reductase, is required for chlorophyll synthesis and plant growth development. Gene 627, 351-362. |
| [41] | Qiu J, Sun SQ, Luo SQ, Zhang JC, Xiao XZ, Zhang LQ, Wang F, Liu SZ (2014). Arabidopsis AtPAP1 transcription factor induces anthocyanin production in transgenic Taraxacum brevicorniculatum. Plant Cell Rep 33, 669-680. |
| [42] | Sakowska K, Alberti G, Genesio L, Peressotti A, Delle Vedove G, Gianelle D, Colombo R, Rodeghiero M, Panigada C, Juszczak R, Celesti M, Rossini M, Haworth M, Campbell BW, Mevy JP, Vescovo L, Cendrero-Mateo MP, Rascher U, Miglietta F (2018). Leaf and canopy photosynthesis of a chlorophyll deficient soybean mutant. Plant Cell Environ 41, 1427-1437. |
| [43] |
Sandhu D, Atkinson T, Noll A, Johnson C, Espinosa K, Boelter J, Abel S, Dhatt BK, Barta T, Singsaas E, Sepsenwol S, Goggi AS, Palmer RG (2016). Soybean proteins GmTic110 and GmPsbP are crucial for chloroplast development and function. Plant Sci 252, 76-87.
DOI PMID |
| [44] |
Schmittgen TD, Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101-1108.
DOI PMID |
| [45] | Shim KC, Kang YN, Song JH, Kim YJ, Kim JK, Kim C, Tai TH, Park I, Ahn SN (2023). A frameshift mutation in the Mg-chelatase I subunit gene OsCHLI is associated with a lethal chlorophyll-deficient, yellow seedling phenotype in rice. Plants 12, 2831. |
| [46] | Song MF, Wei QZ, Wang J, Fu WY, Qin XD, Lu XM, Cheng F, Yang K, Zhang L, Yu XQ, Li J, Chen JF, Lou QF (2018). Fine mapping of CsVYL, conferring virescent leaf through the regulation of chloroplast development in cucumber. Front Plant Sci 9, 432. |
| [47] | Stern DB, Hanson MR, Barkan A (2004). Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci 9, 293-301. |
| [48] | Sun Y, Bai PP, Gu KJ, Yang SZ, Lin HY, Shi CG, Zhao YP (2022). Dynamic transcriptome and network-based analysis of yellow leaf mutant Ginkgo biloba. BMC Plant Biol 22, 465. |
| [49] | Tanaka R, Tanaka A (2011). Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim Biophys Acta Bioenerg 1807, 968-976. |
| [50] |
Wu ZM, Zhang X, He B, Diao LP, Sheng SL, Wang JL, Guo XP, Su N, Wang LF, Jiang L, Wang CM, Zhai HQ, Wan JM (2007). A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145, 29-40.
DOI PMID |
| [51] |
Xie Y, Tan HJ, Ma ZX, Huang JR (2016). DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol Plant 9, 711-721.
DOI PMID |
| [52] | Xiong LR, Du H, Zhang KY, Lv D, He HL, Pan JS, Cai R, Wang G (2021). A mutation in CsYL2.1 encoding a plastid isoform of triose phosphate isomerase leads to yellow leaf 2.1 (yl2.1) in cucumber (Cucumis sativus L.). Int J Mol Sci 22, 322. |
| [53] |
Xu BH, Zhang CY, Gu Y, Cheng R, Huang DY, Liu X, Sun YD (2023). Physiological and transcriptomic analysis of a yellow leaf mutant in watermelon. Sci Rep 13, 9647.
DOI PMID |
| [54] |
Yin JJ, Zhu XB, Yuan C, Wang J, Li WT, Wang YP, He M, Cheng QS, Ye BQ, Chen WL, Linghu QY, Wang JC, Ma BT, Qin P, Li SG, Chen XW (2015). Characterization and fine mapping of a novel vegetative senescence lethal mutant locus in rice. J Genet Genomics 42, 511-514.
DOI PMID |
| [55] | Zhang HT, Li JJ, Yoo JH, Yoo SC, Cho SH, Koh HJ, Seo HS, Paek NC (2006). Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 62, 325-337. |
| [56] | Zhang KJ, Li Y, Zhu WW, Wei YF, Njogu MK, Lou QF, Li J, Chen JF (2020b). Fine mapping and transcriptome analysis of virescent leaf gene v-2 in cucumber (Cucumis sativus L.). Front Plant Sci 11, 570817. |
| [57] | Zhang TT, Dong XY, Yuan X, Hong YY, Zhang LL, Zhang X, Chen SX (2022). Identification and characterization of CsSRP43, a major gene controlling leaf yellowing in cucumber. Hortic Res 9, uhac212. |
| [58] | Zhao CJ, Liu LJ, Safdar LB, Xie ML, Cheng XH, Liu YY, Xiang Y, Tong CB, Tu JX, Huang JY, Liu SY (2020a). Characterization and fine mapping of a yellow-virescent gene regulating chlorophyll biosynthesis and early stage chloroplast development in Brassica napus. G3(Bethesda) 10, 3201-3211. |
| [59] | Zhong XM, Sun SF, Li FH, Wang J, Shi ZS (2015). Photosynthesis of a yellow-green mutant line in maize. Photosynthetica 53, 499-505. |
| [60] | Zhou H, Lin-Wang K, Wang HL, Gu C, Dare AP, Espley RV, He HP, Allan AC, Han YP (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J 82, 105-121. |
| [61] |
Zhu HY, Zhang MJ, Sun SR, Yang S, Li JX, Li H, Yang HH, Zhang KG, Hu JB, Liu DM, Yang LM (2019). A single nucleotide deletion in an ABC transporter gene leads to a dwarf phenotype in watermelon. Front Plant Sci 10, 1399.
DOI PMID |
| [62] | Zhu XY, Guo S, Wang ZW, Du Q, Xing YD, Zhang TQ, Shen WQ, Sang XC, Ling YH, He GH (2016). Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa). BMC Plant Biol 16, 134. |
| [1] | Binqi Li, Jiahui Yan, Hao Li, Wei Xin, Yunhe Tian, Zhenbiao Yang, Wenxin Tang. Changes of Small GTPases Activity During Cucumber Tendril Winding [J]. Chinese Bulletin of Botany, 2022, 57(3): 299-307. |
| [2] | Zeyi Wang, Hengjia Zhang, Yucai Wang, Xietian Chen, Yuchun Ba. Effects of Deficit Irrigation on the Photosynthetic and Physiological Characteristics of Leaves and Yield of Isatis tinctoria [J]. Chinese Bulletin of Botany, 2020, 55(6): 705-714. |
| [3] | Jianfu Liu, Yucai Chen, Wenjian Wang, Hechuan Wang, Jinfu Cai, Mingyuan Wang, Dandan Li, Bin Zhang, Kun Huang. Effects of Space Treatment on Biological and Growth Characteristics of Camellia sinensis [J]. Chinese Bulletin of Botany, 2020, 55(5): 564-572. |
| [4] | Dongdong Cao,Shanyu Chen,Yebo Qin,Huaping Wu,Guanhai Ruan,Yutao Huang. Regulatory Mechanism of Salicylic Acid on Seed Germination Under Salt Stress in Kale [J]. Chinese Bulletin of Botany, 2020, 55(1): 49-61. |
| [5] | Chun Zhou,Ran Jiao,Ping Hu,Han Lin,Juan Hu,Na Xu,Xianmei Wu,Yuchun Rao,Yuexing Wang. Gene Mapping and Candidate Gene Analysis of Rice Early Senescence Mutant LS-es1 [J]. Chinese Bulletin of Botany, 2019, 54(5): 606-619. |
| [6] | Amangul·Mambetale, Lazati·Nurbulat, Lili Gao, Jusong Zhang, Liwen Tian. Effect of Salt Stress on Growth and Physiological Characteristics of Sea Island Cotton and Upland Cotton Cultivars [J]. Chinese Bulletin of Botany, 2017, 52(4): 465-473. |
| [7] | Hu Chaoqin, , Liu Jianyu, , Wang Yunqian, Yang Rui, Wang Bingkun, He Yueqiu, Zeng Qianchun, Luo Qiong. Mapping of Pizy6(t), a Gene Conferring Resistance to the Rice Blast Strain LP11, in Oryza sativa subsp. japonica Cultivar Ziyu44 [J]. Chinese Bulletin of Botany, 2017, 52(1): 61-69. |
| [8] | Wei Wang, Jiayu Wang, Shenglong Yang, Jin Liu, Xiaoyan Dong, Guojiao Wang, Wenfu Chen. Identification and Gene Mapping of the nrl7 Mutant in Rice [J]. Chinese Bulletin of Botany, 2016, 51(3): 290-295. |
| [9] | Lei Deng, Minmin Du, Chuanyou Li. Chinese Scientists Made Breakthrough Progresses in Studies on Domestication and Fruit Quality in Tomato and Cucumber [J]. Chinese Bulletin of Botany, 2015, 50(3): 275-278. |
| [10] | Dan Liu, Jiayu Wang, Jin Liu, Dianrong Ma, Minghui Zhao, Wenfu Chen. Genetic Analysis and Gene Mapping of a Rice Spreadingpanicle Mutant [J]. Chinese Bulletin of Botany, 2015, 50(2): 198-205. |
| [11] | Yudong Jiang, Peilong He, Hongxiang Liao, Xiaobo Zhang, Guochao Wu, Guanghua He, Tingting Lin, Xianchun Sang. Identification and Gene Mapping of a Fragile and Leaf-tip Dead Mutant fld1 in Oryza sativa [J]. Chinese Bulletin of Botany, 2014, 49(6): 663-671. |
| [12] | Ye Wang, Xingfang Gu, Shengping Zhang, Han Miao, Guohua Chen, Bingyan Xie. Transformation of RNAi Vector in Cucumber (Cucumis sativus) In Vitro by Agrobacterium tumefaciens-mediated Transfection [J]. Chinese Bulletin of Botany, 2014, 49(2): 183-189. |
| [13] | Huanhuan Wang, Qiang Cai, Mingjiao Chen, Zhijing Luo, Dabing Zhang, Zheng Yuan. Phenotype Analyses and Gene Mapping of abnormal palea and lodicules, a Rice Mutant with Abnormal Floral Organs [J]. Chinese Bulletin of Botany, 2014, 49(1): 1-7. |
| [14] | Yongdong Sun, Weirong Luo, Xinzheng Li, Guangyin Wang. Effect of Exogenous NaHS on Seed Germination and Physiological Characteristics of Cucurbita ficifolia Under NaHCO3 Stress [J]. Chinese Bulletin of Botany, 2014, 49(1): 98-104. |
| [15] | Dewei Yang, Meijuan Zeng, Libin Lu, Ning Ye, Chengde Liu, Xianghua Zheng, Xinfu Ye. Genetic Analysis and Mapping of Rice Dwarf Mutant ds1 [J]. Chinese Bulletin of Botany, 2011, 46(6): 617-624. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||