

Chinese Bulletin of Botany ›› 2021, Vol. 56 ›› Issue (1): 1-5.DOI: 10.11983/CBB20208 cstr: 32102.14.CBB20208
• COMMENTARIES • Next Articles
Received:2020-12-25
Accepted:2020-12-29
Online:2021-01-01
Published:2021-01-15
Contact:
Guohua Xu
Wei Xuan, Guohua Xu. Genomic Basis of Rice Adaptation to Soil Nitrogen Status[J]. Chinese Bulletin of Botany, 2021, 56(1): 1-5.
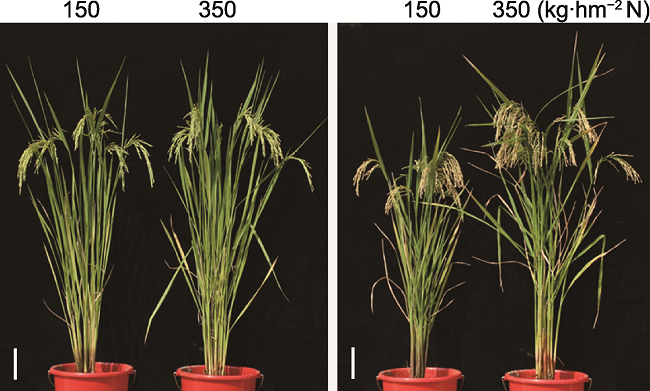
Figure 1 Rice landraces from different geographical regions show different sensitivity to soil nitrogen content The indica rice variety S23 (left panel) and variety Y5 (right panel) are insensitive and sensitive to the change of soil nitrogen content, respectively (provided by Guohua Xu’s lab and photographed by Le Luo). Bars=10 cm
| [1] |
Fan XR, Tang Z, Tan YW, Zhang Y, Luo BB, Yang M, Lian XM, Shen QR, Miller AJ, Xu GH (2016). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc Natl Acad Sci USA 113, 7118-7123.
DOI URL PMID |
| [2] |
Gao ZY, Wang YF, Chen G, Zhang AP, Yang SL, Shang LG, Wang DY, Ruan BP, Liu CL, Jiang HZ, Dong GJ, Zhu L, Hu J, Zhang GH, Zeng DL, Guo LB, Xu GH, Teng S, Harberd NP, Qian Q (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat Commun 10, 5207.
DOI URL PMID |
| [3] | Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, Zhang WF, Christie P, Goulding KWT, Vitousek PM, Zhang FS (2010). Significant acidification in major Chinese crop-lands. Science 327, 1008-1010. |
| [4] | Hu B, Wang W, Ou SJ, Tang JY, Li H, Che RH, Zhang ZH, Chai XY, Wang HR, Wang YQ, Liang CZ, Liu LC, Piao ZZ, Deng QY, Deng K, Xu C, Liang Y, Zhang LH, Li LG, Chu CC (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47, 834-838. |
| [5] | Li S, Tian YH, Wu K, Ye YF, Yu JP, Zhang JQ, Liu Q, Hu MY, Li H, Tong YP, Harberd NP, Fu XD (2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595-600. |
| [6] |
Liu YQ, Wang HR, Jiang ZM, Wang W, Xu RN, Wang QH, Zhang ZH, Li AF, Liang Y, Ou SJ, Liu XJ, Cao SY, Tong HN, Wang YH, Zhou F, Liao H, Hu B, Chu CC (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature https://doi.org/10.1038/s41586-020-03091-w.
URL PMID |
| [7] |
Stevens CJ (2019). Nitrogen in the environment. Science 363, 578-580.
URL PMID |
| [8] |
Sun HY, Qian Q, Wu K, Luo JJ, Wang SS, Zhang CW, Ma YF, Liu Q, Huang XZ, Yuan QB, Han RX, Zhao M, Dong GJ, Guo LB, Zhu XD, Gou ZH, Wang W, Wu YJ, Lin HX, Fu XD (2014). Heterotrimeric G protein regulate nitrogen-use efficiency in rice. Nat Genet 46, 652-656.
URL PMID |
| [9] | Tang WJ, Ye J, Yao XM, Zhao PZ, Xuan W, Tian YL, Zhang YY, Xu S, An HZ, Chen GM, Yu J, Wu W, Ge YW, Liu XL, Li J, Zhang HZ, Zhao YQ, Yang B, Jiang XZ, Peng C, Zhou C, Terzaghi W, Wang CM, Wan JM (2019). Genome-wide associated study identifies NAC42- activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat Commun 10, 5279. |
| [10] | Wang Q, Nian JQ, Xie XZ, Yu H, Zhang J, Bai JT, Dong GJ, Hu J, Bai B, Chen LC, Xie QJ, Feng J, Yang XL, Peng JL, Chen F, Qian Q, Li JY, Zuo J (2018). Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat Commun 9, 735. |
| [11] | Wang SS, Chen AQ, Xie K, Yang XF, Luo ZZ, Chen JD, Zeng DC, Ren YH, Yang CF, Wang LX, Feng HM, López-Arredondo DL, Herrera-Estrella LR, Xu GH (2020). Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc Natl Acad Sci USA 117, 16649-16659. |
| [12] | Wu K, Wang SS, Song WZ, Zhang JQ, Wang Y, Liu Q, Yu JP, Ye YF, Li S, Chen JF, Zhao Y, Wang J, Wu XK, Wang MY, Zhang YJ, Liu BM, Wu YJ, Harberd NP, Fu XD (2020). Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367, eaaz2046. |
| [13] | Xu GH, Fan XR, Miller AJ (2012). Plant nitrogen assimilation and use efficiency. Ann Rev Plant Biol 63, 153-182. |
| [14] | Zhang JY, Liu YX, Zhang N, Hu B, Jin T, Xu HR, Qin Y, Yan PX, Zhang XN, Guo XX, Hui J, Cao SY, Wang X, Wang C, Wang H, Qu BY, Fan GY, Yuan LX, Garrido-Oter R, Chu CC, Bai Y (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol 37, 676-684. |
| [15] |
Zhang S, Zhang Y, Li K, Yan M, Zhang JF, Yu M, Tang S, Wang LY, Qu HY, Luo L, Xuan W, Xu GH (2020). Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr Biol 31, 1-13.
DOI URL PMID |
| [1] | ZHAO Hong-Xian, LIU Peng, SHI Man-Ying, XU Ming-Ze, JIA Xin, TIAN Yun, ZHA Tian-Shan. Effect of leaf nitrogen allocation on maximum net photosynthetic rate of two common sand-fixing species, Artemisia ordosica and Leymus secalinus, in Mau Us Sandy Land [J]. Chin J Plant Ecol, 2025, 49(3): 460-474. |
| [2] | GENG Xue-Qi, TANG Ya-Kun, WANG Li-Na, DENG Xu, ZHANG Ze-Ling, ZHOU Ying. Nitrogen addition increases biomass but reduces nitrogen use efficiency of terrestrial plants in China [J]. Chin J Plant Ecol, 2024, 48(2): 147-157. |
| [3] | Bangbang Wu, Yuqiong Hao, Shubin Yang, Yuxi Huang, Panfeng Guan, Xingwei Zheng, Jiajia Zhao, Ling Qiao, Xiaohua Li, Weizhong Liu, Jun Zheng. Evaluation and Genetic Variation of Grain Lutein Contents in Common Wheat From Shanxi [J]. Chinese Bulletin of Botany, 2023, 58(4): 535-547. |
| [4] | FENG Xu-Fei, LEI Zhang-Ying, ZHANG Yu-Jie, XIANG Dao, YANG Ming-Feng, ZHANG Wang-Feng, ZHANG Ya-Li. Effect of leaf nitrogen allocation on photosynthetic nitrogen use efficiency at flowering and boll stage of Gossypium spp. [J]. Chin J Plant Ecol, 2023, 47(11): 1600-1610. |
| [5] | Yuan Li, Yujie Chang, Lanfen Wang, Shumin Wang, Jing Wu. Screening of Resistance Germplasm Resources and Genome-wide Association Study of Fusarium Wilt in Common Bean [J]. Chinese Bulletin of Botany, 2023, 58(1): 51-61. |
| [6] | Liu Xiaolong, Ji Ping, Yang Hongtao, Ding Yongdian, Fu Jialing, Liang Jiangxia, Yu Congcong. Priming Effect of Abscisic Acid on High Temperature Stress During Rice Heading-flowering Stage [J]. Chinese Bulletin of Botany, 2022, 57(5): 596-610. |
| [7] | Jiaxin Li, Xia Li, Yinfeng Xie. Mechanism on Drought Tolerance Enhanced by Exogenous Trehalose in C4-PEPC Rice [J]. Chinese Bulletin of Botany, 2021, 56(3): 296-314. |
| [8] | Yuhui Zhao, Xiuxiu Li, Zhuo Chen, Hongwei Lu, Yucheng Liu, Zhifang Zhang, Chengzhi Liang. An Overview of Genome-wide Association Studies in Plants [J]. Chinese Bulletin of Botany, 2020, 55(6): 715-732. |
| [9] | Ningxi Song, Yingfeng Xie, Xia Li. Effects of Epigenetic Mechanisms on C4 Phosphoenolpyruvate Carboxylase Transgenic Rice (Oryza sativa) Seed Germination Under Drought Stress [J]. Chinese Bulletin of Botany, 2020, 55(6): 677-692. |
| [10] | Lu Zhang,Xinhua He. Nitrogen Utilization Mechanism in C3 and C4 Plants [J]. Chinese Bulletin of Botany, 2020, 55(2): 228-239. |
| [11] | Mei-ling Han,Ru-jiao Tan,Dai-yin Chao. A New Progress of Green Revolution: Epigenetic Modification Dual-regulated by Gibberellin and Nitrogen Supply Contributes to Breeding of High Yield and Nitrogen Use Efficiency Rice [J]. Chinese Bulletin of Botany, 2020, 55(1): 5-8. |
| [12] | FU Yi-Wen, TIAN Da-Shuan, WANG Jin-Song, NIU Shu-Li, ZHAO Ken-Tian. Patterns and affecting factors of nitrogen use efficiency of plant leaves and roots in Nei Mongol and Qinghai-Xizang Plateau grasslands [J]. Chin J Plant Ecol, 2019, 43(7): 566-575. |
| [13] | Lin Chen,Yan Lin,Pengfei Chen,Shaohua Wang,Yanfeng Ding. Effect of Iron Deficiency on the Protein Profile of Rice (Oryza sativa) Phloem Sap [J]. Chinese Bulletin of Botany, 2019, 54(2): 194-207. |
| [14] | LI Xin-Hao, YAN Hui-Juan, WEI Teng-Zhou, ZHOU Wen-Jun, JIA Xin, ZHA Tian-Shan. Relative changes of resource use efficiencies and their responses to environmental factors in Artemisia ordosica during growing season [J]. Chin J Plant Ecol, 2019, 43(10): 889-898. |
| [15] | ZHU Qi-Lin, XIANG Rui, TANG Li, LONG Guang-Qiang. Effects of intercropping on photosynthetic rate and net photosynthetic nitrogen use efficiency of maize under nitrogen addition [J]. Chin J Plan Ecolo, 2018, 42(6): 672-680. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
