

Chinese Bulletin of Botany ›› 2020, Vol. 55 ›› Issue (3): 299-307.DOI: 10.11983/CBB19217 cstr: 32102.14.CBB19217
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Huijin Fan1,2,Kangming Jin1,Renying Zhuo1,Guirong Qiao1,*( )
)
Received:2019-11-07
Accepted:2020-03-24
Online:2020-05-01
Published:2020-07-06
Contact:
Guirong Qiao
Huijin Fan, Kangming Jin, Renying Zhuo, Guirong Qiao. Cloning and Expression Analysis of Different Truncated U3 Promoters in Phyllostachys edulis[J]. Chinese Bulletin of Botany, 2020, 55(3): 299-307.
| Primer name | Sequences (5'-3') |
|---|---|
| PeU3-1 | F: ATCCGCCCCTGGTTGCTCTA R: CAAGCCGTTAATCACGCTCTGG |
| PeU3-2 | F: TCAGCGTTGACCTCCTCTTG R: TGGCACGAAATGGGATGAAG |
| 1301-PeU3-1-1-F | F: GACCTGCAGGCATGCAAGCTTTCAGCGTTGACCTCCTCTT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-1-2-F | F: GACCTGCAGGCATGCAAGCTCAGGTCATAAGATGGAAGCT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-1-3-F | F: GACCTGCAGGCATGCAAGCTCAGCCCATACGAAAGTGATT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-2-1-F | F: GACCTGCAGGCATGCAAGCTCCCTAGTGATGCCTTAAAGC R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| 1301-PeU3-2-2-F | F: GACCTGCAGGCATGCAAGCTATGGTATTCTGTTGCGGACT R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| 1301-PeU3-2-3-F | F: GACCTGCAGGCATGCAAGCTTGGCCTTTTTCATGGAGCCG R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-1-1-F | F: TCGACGGTATCGATAAGCTTTTCAGCGTTGACCTCCTCTT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-1-2-F | F: TCGACGGTATCGATAAGCTTCAGGTCATAAGATGGAAGCT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-1-3-F | F: TCGACGGTATCGATAAGCTTCAGCCCATACGAAAGTGATT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-2-1-F | F: TCGACGGTATCGATAAGCTTATCCGCCCCTGGTTGCTCTA R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-2-2-F | F: TCGACGGTATCGATAAGCTTATGGTATTCTGTTGCGGACT R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-2-3-F | F: TCGACGGTATCGATAAGCTTTGGCCTTTTTCATGGAGCCG R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
Table 1 Primer sequences used in this study
| Primer name | Sequences (5'-3') |
|---|---|
| PeU3-1 | F: ATCCGCCCCTGGTTGCTCTA R: CAAGCCGTTAATCACGCTCTGG |
| PeU3-2 | F: TCAGCGTTGACCTCCTCTTG R: TGGCACGAAATGGGATGAAG |
| 1301-PeU3-1-1-F | F: GACCTGCAGGCATGCAAGCTTTCAGCGTTGACCTCCTCTT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-1-2-F | F: GACCTGCAGGCATGCAAGCTCAGGTCATAAGATGGAAGCT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-1-3-F | F: GACCTGCAGGCATGCAAGCTCAGCCCATACGAAAGTGATT R: TTTACCCTCAGATCTACCATTGCTGAGTAGCGCAGGGCTC |
| 1301-PeU3-2-1-F | F: GACCTGCAGGCATGCAAGCTCCCTAGTGATGCCTTAAAGC R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| 1301-PeU3-2-2-F | F: GACCTGCAGGCATGCAAGCTATGGTATTCTGTTGCGGACT R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| 1301-PeU3-2-3-F | F: GACCTGCAGGCATGCAAGCTTGGCCTTTTTCATGGAGCCG R: TTTACCCTCAGATCTACCATTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-1-1-F | F: TCGACGGTATCGATAAGCTTTTCAGCGTTGACCTCCTCTT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-1-2-F | F: TCGACGGTATCGATAAGCTTCAGGTCATAAGATGGAAGCT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-1-3-F | F: TCGACGGTATCGATAAGCTTCAGCCCATACGAAAGTGATT R: GCTCTAGAACTAGTGGATCCTGCTGAGTAGCGCAGGGCTC |
| LUC-PeU3-2-1-F | F: TCGACGGTATCGATAAGCTTATCCGCCCCTGGTTGCTCTA R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-2-2-F | F: TCGACGGTATCGATAAGCTTATGGTATTCTGTTGCGGACT R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
| LUC-PeU3-2-3-F | F: TCGACGGTATCGATAAGCTTTGGCCTTTTTCATGGAGCCG R: GCTCTAGAACTAGTGGATCCTGCATCAATGCTCCAAAGCT |
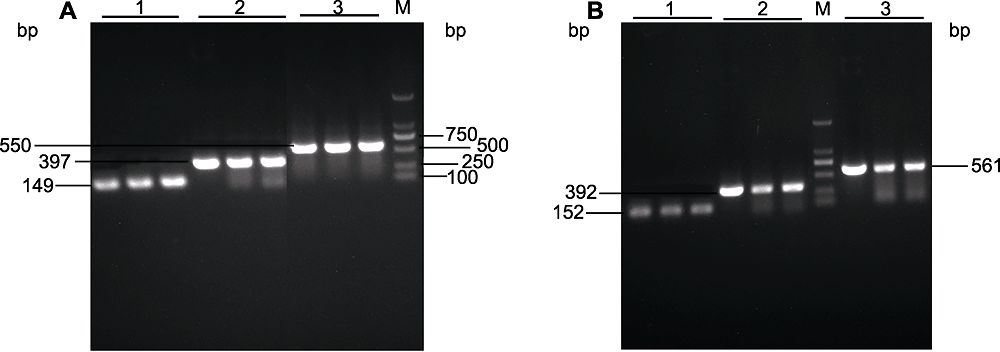
Figure 2 PCR analysis of six truncated PeU3 promoters (A) Amplification using PeU3-1 promoter plasmid as template (1: PeU3-1-3pro; 2: PeU3-1-2pro; 3: PeU3-1-1pro); (B) Amplification using PeU3-2 promoter plasmid as template (1: PeU3-2-3pro; 2: PeU3-2-2pro; 3: PeU3-2-1pro). M: 2 kb DNA marker

Figure 4 GUS expression driven by different truncated PeU3 promoters (A) Transient expression of GUS gene driven by different truncated PeU3 promoters in the leaves of Nicotiana benthamiana (Bars=5 mm); (B) Expression of GUS gene driven by different truncated PeU3 promoters in the callus of Dendrocalamus latiflorus (Bars=5 mm). CK: Negative control
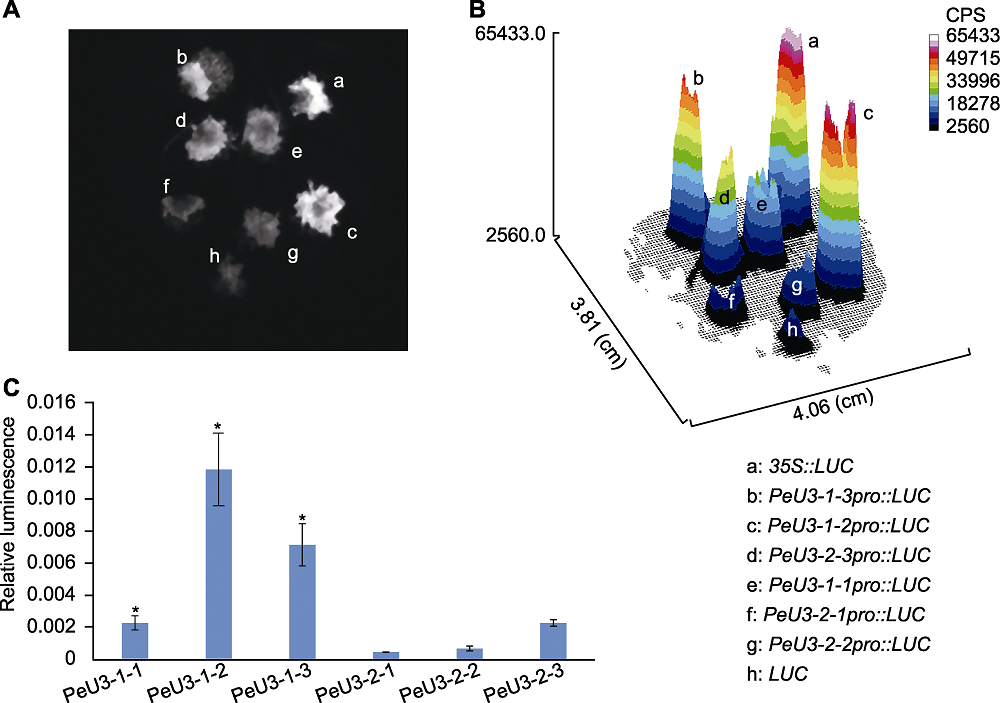
Figure 5 LUC expression driven by different truncated PeU3 promoters in the leaves of Nicotiana benthamiana (A), (B) Transient expression LUC assays illustrating the activation of PeU3 promoter in leaves of N. benthamiana; (C) The quantification of the relative luminescence intensities (In each experiment, four biological replications were performed with quanti?cation. Asterisk above the bars denote significant differences determined by the Duncan’s test, P<0.05); CPS: Luminescence intensity
| [1] | 陈香嵩, 李甜甜, 周少立, 赵毓 (2018). 外源蛋白在烟草叶片瞬时表达. BioProtoc doi: 10.217691BioProtoc.1010127. |
| [2] | 雷建峰, 李月, 徐新霞, 阿尔祖古丽·塔什, 蒲艳, 张巨松, 刘晓东 (2016a). 棉花不同GbU6启动子截短克隆及功能鉴定. 作物学报 42, 675-683. |
| [3] | 雷建峰, 徐新霞, 代培红, 李继洋, 张巨松, 刘晓东 (2016b). 不同截短U3启动子在棉花中的功能分析. 棉花学报 28, 307-314. |
| [4] | 李继洋, 雷建峰, 代培红, 姚瑞, 曲延英, 陈全家, 李月, 刘晓东 (2018). 基于棉花U6启动子的海岛棉CRISPR/Cas9基因组编辑体系的建立. 作物学报 44, 227-235. |
| [5] | 李丽莉, 扈廷茂, 扈会平, 刘明秋, 苏慧敏 (2005). 利用番茄U3snRNA基因上游启动区构建植物表达载体及对烟草的转化. 内蒙古大学学报(自然科学版) 36, 63-67. |
| [6] | 蒲艳, 刘晓东, 阿尔祖古丽·塔什, 魏倩, 刘超 (2019). 番茄不同截短U3启动子的克隆及功能分析. 华北农学报 34, 33-39. |
| [7] | 孙建飞, 翟建云, 马元丹, 傅卢成, 卜柯丽, 王柯杨, 高岩, 张汝民 (2018). 毛竹快速生长期茎秆不同节间光合色素和光合酶活性的差异. 植物学报 53, 773-781. |
| [8] | 藏旭阳, 代培红, 李继洋, 蒲艳, 顾爱星, 刘晓东 (2019). 棉花U3和U6启动子在CRISPR/Cas9基因组编辑体系中的功能鉴定. 棉花学报 31, 31-39. |
| [9] | Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug II RG, Tan WF, Penheiter SG, Ma AC, Leung AYH, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114-118. |
| [10] | Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, 39. |
| [11] |
Bibikova M, Beumer K, Trautman JK, Carroll D (2003). Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764-764.
URL PMID |
| [12] | Bruegmann T, Fladung M (2019). Overexpression of both flowering time genes AtSOC1 and SaFUL revealed huge influence onto plant habitus in poplar. Tree Genet Genomes 15, 20. |
| [13] | Charrier A, Vergen E, Dousset N, Richer A, Petiteau A, Chevreau E (2019). Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR- Cas9 system. Front Plant Sci 10, 40. |
| [14] | Chen JR, Shafi M, Li S, Wang J, Wu JS, Ye ZQ, Peng DL, Yan WB, Liu D (2015). Copper induced oxidative stresses, antioxidant responses and phytoremediation potential of Moso bamboo (Phyllostachys pubescens). Sci Rep 5, 13554. |
| [15] | Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. |
| [16] | Doll NM, Gilles LM, Gérentes MF, Richard C, Just J, Fierlej Y, Borrelli VMG, Gendrot G, Ingram GC, Rogowsky PM, Widiez T (2019). Single and multiple gene knockouts by CRISPR-Cas9 in maize. Plant Cell Rep 38, 487-501. |
| [17] |
Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67-71.
DOI URL PMID |
| [18] |
Ge W, Zhang Y, Cheng ZC, Hou D, Li XP, Gao J (2017). Main regulatory pathways, key genes and microRNAs involved in flower formation and development of Moso bamboo (Phyllostachys edulis). Plant Biotechnol J 15, 82-96.
URL PMID |
| [19] |
Jinek M, Chylinski K, Fonfara L, Hauer M, Doudna JA, Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821.
URL PMID |
| [20] | Lee K, Zhang YX, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK, Qi YP, Wang K (2019). Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J 17, 362-372. |
| [21] |
Li L, Cheng ZC, Ma YJ, Bai QS, Li XY, Cao ZH, Wu ZN, Gao J (2018). The association of hormone signaling genes, transcription and changes in shoot anatomy during Moso bamboo growth. Plant Biotechnol J 16, 72-85.
DOI URL PMID |
| [22] | Li X, Jiang DH, Yong KL, Zhang DB (2007). Varied transcriptional efficiencies of multiple Arabidopsis U6 small nuclear RNA genes. J Integr Plant Biol 49, 222-229. |
| [23] | Liang Z, Chen KL, Li TD, Zhang Y, Wang YP, Zhao Q, Liu JX, Zhang HW, Liu CM, Ran YD, Gao CX (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8, 14261. |
| [24] |
Liang Z, Zhang K, Chen KL, Gao CX (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/ Cas system. J Genet Genomics 41, 63-68.
URL PMID |
| [25] |
Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, Xie YY, Shen RX, Chen SF, Wang Z, Chen YL, Guo JX, Chen LT, Zhao XC, Dong ZC, Liu YG (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8, 1274-1284.
URL PMID |
| [26] |
Marshallsay C, Connelly S, Filipowicz W (1992). Characterization of the U3 and U6 snRNA genes from wheat: U3 snRNA genes in monocot plants are transcribed by RNA polymerase III. Plant Mol Biol 19, 973-983.
URL PMID |
| [27] |
Nandy S, Pathak B, Zhao S, Srivastava V (2019). Heat- shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct 3, e00145.
DOI URL PMID |
| [28] |
Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 691-693.
URL PMID |
| [29] |
Papikian A, Liu WL, Gallego-Bartolomé J, Jacobsen SE (2019). Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun 10, 729.
URL PMID |
| [30] |
Peng ZH, Lu Y, Li LB, Zhao Q, Feng Q, Gao ZM, Lu HY, Hu T, Yao N, Liu KY, Li Y, Fan DL, Guo YL, Li WJ, Lu YQ, Weng QJ, Zhou CC, Zhang L, Huang T, Zhao Y, Zhu CR, Liu XG, Yang XW, Wang T, Miao K, Zhuang CY, Cao XL, Tang WL, Liu GS, Liu YL, Chen J, Liu ZJ, Yuan LC, Liu ZH, Huang XH, Lu TT, Fei BH, Ning ZM, Han B, Jiang ZH (2013). The draft genome of the fast-growing non-timber forest species Moso bamboo (Phyllostachys heterocycla). Nat Genet 45, 456-461.
DOI URL PMID |
| [31] |
Ren C, Guo YC, Gathunga EK, Duan W, Li SH, Liang ZC (2019). Recovery of the non-functional EGFP-assisted identification of mutants generated by CRISPR/Cas9. Plant Cell Rep 38, 1541-1549.
DOI URL PMID |
| [32] | Sandhu M, Wani SH, Jiménez VM (2018). In vitro propagation of bamboo species through axillary shoot proliferation: a review. Plant Cell Tissue Organ Cult 132, 27-53. |
| [33] |
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T (2014). CRISPR/Cas9- mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55, 475-481.
URL PMID |
| [34] |
Wang MB, Helliwell CA, Wu LM, Waterhouse PM, Peacock WJ, Dennis ES (2008). Hairpin RNAs derived from RNA polymerase II and polymerase III promoter-directed transgenes are processed differently in plants. RNA 14, 903-913.
URL PMID |
| [35] | Ye SW, Chen G, Kohnen MV, Wang WJ, Cai CY, Ding WS, Wu C, Gu LF, Zheng YS, Ma XQ, Lin CT, Zhu Q (2020). Robust CRISPR/Cas9 mediated genome editing and its application in manipulating plant height in the first generation of hexaploid Ma bamboo (Dendrocalamus latiflorus Munro). Plant Biotechnol J doi: 10.1111/pbi.13320. |
| [36] |
Zhang JS, Zhang H, Botella JR, Zhu JK (2018). Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol 60, 369-375.
DOI URL PMID |
| [37] |
Zhang ZZ, Hua L, Gupta A, Tricoli D, Edwards KJ, Yang B, Li WL (2019). Development of an Agrobacterium- delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol J 17, 1623-1635.
URL PMID |
| [38] | Zhao HS, Gao ZM, Wang L, Wang JL, Wang SB, Fei BH, Chen CH, Shi CC, Liu XC, Zhang HL, Lou YF, Chen LF, Sun HY, Zhou XQ, Wang SN, Zhang C, Xu H, Li LC, Yang YH, Wei YL, Yang W, Gao Q, Yang HM, Zhao SC, Jiang ZH (2018). Chromosome-level reference genome and alternative splicing atlas of Moso bamboo (Phyllostachys edulis). Gigascience 7, giy115. |
| [1] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [2] | Danling Hu, Yongwei Sun. Advances in Virus-mediated Genome Editing Technology in Plants [J]. Chinese Bulletin of Botany, 2024, 59(3): 452-462. |
| [3] | Na Wang, Teng Jiang, Binxi Wang, Lifang Niu, Hao Lin. Advances in Haploid Breeding Technology and Its Application in Alfalfa and Other Legume Forages [J]. Chinese Bulletin of Botany, 2022, 57(6): 756-763. |
| [4] | Lubin Tan, Chuanqing Sun. Rapid Domestication of Wild Allotetraploid Rice: Starting a New Era of Human Agricultural Civilization [J]. Chinese Bulletin of Botany, 2021, 56(2): 134-137. |
| [5] | Xianrong Xie, Dongchang Zeng, Jiantao Tan, Qinlong Zhu, Yaoguang Liu. CRISPR-based DNA Fragment Deletion in Plants [J]. Chinese Bulletin of Botany, 2021, 56(1): 44-49. |
| [6] | Yuekai Su,Jingren Qiu,Han Zhang,Zhenqiao Song,Jianhua Wang. Recent Progress in Evolutionary Technology of CRISPR/Cas9 System for Plant Genome Editing [J]. Chinese Bulletin of Botany, 2019, 54(3): 385-395. |
| [7] | Sun Jianfei, Zhai Jianyun, Ma Yuandan, , Fu Lucheng, Bu Keli, Wang Keyang, Gao Yan, Zhang Rumin. Differences in Photosynthetic Pigments and Photosynthetic Enzyme Activities in Different Internodes of Phyllostachys edulis During Rapid Growth Stage [J]. Chinese Bulletin of Botany, 2018, 53(6): 773-781. |
| [8] | Rui-Yu ZHAO, Zheng-Cai LI, Bin WANG, Xiao-Gai GE, Yun-Xi DAI, Zhi-Xia ZHAO, Yu-Jie ZHANG. Duration of mulching caused variable pools of labile organic carbon in a Phyllostachys edulis plantation [J]. Chin J Plant Ecol, 2017, 41(4): 418-429. |
| [9] | Xiao-Gai GE, Ben-Zhi ZHOU, Wen-Fa XIAO, Xiao-Ming WANG, Yong-Hui CAO, Ming YE. Effects of biochar addition on dynamics of soil respiration and temperature sensitivity in a Phyllostachys edulis forest [J]. Chin J Plant Ecol, 2017, 41(11): 1177-1189. |
| [10] | Ming Ouyang, Qingpei Yang, Xin Chen, Guangyao Yang, Jianmin Shi, Xiangmin Fang. Effects of the expansion of Phyllostachys edulis on species composition, structure and diversity of the secondary evergreen broad-leaved forests [J]. Biodiv Sci, 2016, 24(6): 649-657. |
| [11] | CHEN Yong-Gang,TANG Meng-Ping,YANG Chun-Ju,MA Tian-Wu,WANG Li. Spatial analysis of competition in natural Phyllostachys edulis community [J]. Chin J Plan Ecolo, 2015, 39(7): 726-735. |
| [12] | WANG Yi-Kun, JIN Ai-Wu, ZHU Qiang-Gen, QIU Yong-Hua, JI Xin-Liang, ZHANG Si-Hai. Effects of fertilization on the relations of diameter at breast height between different-aged ramets of Phyllostachys edulis population [J]. Chin J Plant Ecol, 2014, 38(3): 289-297. |
| [13] | Yanrong Fan,Shuanglin Chen,Hua Lin,Qingping Yang,Yicong Hong,Ziwu Guo. Effects of different disturbance measures on spatial distribution patterns of understory plants in Phyllostachys edulis forests [J]. Biodiv Sci, 2013, 21(6): 709-714. |
| [14] | Shangbin Bai,Guomo Zhou,Yixiang Wang,Qianqian Liang,Juan Chen,Yanyan Cheng,Rui Shen. Plant species diversity and dynamics in forests invaded by Moso bamboo (Phyllostachys edulis) in Tianmu Mountain Nature Reserve [J]. Biodiv Sci, 2013, 21(3): 288-295. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||