

植物学报 ›› 2022, Vol. 57 ›› Issue (4): 508-531.DOI: 10.11983/CBB22020 cstr: 32102.14.CBB22020
收稿日期:2022-01-21
修回日期:2022-04-24
出版日期:2022-07-01
发布日期:2022-07-14
通讯作者:
陈析丰
作者简介:第一联系人:† 共同第一作者
基金资助:
He Xiaoling, Liu Pengcheng, Ma Bojun, Chen Xifeng( )
)
Received:2022-01-21
Revised:2022-04-24
Online:2022-07-01
Published:2022-07-14
Contact:
Chen Xifeng
About author:* E-mail: xfchen@zjnu.cn摘要: CRISPR/Cas9技术是利用RNA靶向引导Cas9核酸酶对基因组中的目标基因进行编辑的生物技术。近年来, 该技术的多种新型基因编辑器更新迅猛, 编辑效果愈加精细和高效, 在作物定向分子设计育种中展现出巨大的应用前景。该文对CRISPR/Cas9及其相关编辑器的技术原理、编辑效果和应用情况进行综述, 并探讨了该技术在应用中面临的问题、应对措施和发展前景, 旨在为相关领域的科研工作者提供参考。
何晓玲, 刘鹏程, 马伯军, 陈析丰. 基于CRISPR/Cas9的基因编辑技术研究进展及其在植物中的应用. 植物学报, 2022, 57(4): 508-531.
He Xiaoling, Liu Pengcheng, Ma Bojun, Chen Xifeng. Advance in Gene-editing Technology Based on CRISPR/Cas9 and Its Application in Plants. Chinese Bulletin of Botany, 2022, 57(4): 508-531.
| 编辑 技术 | 编辑器 | 脱氨酶/逆转录酶 | Cas蛋白 | PAM (5′-3′) | 活性 窗口 | 编辑类型 | 特点 | 参考文献 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 胞嘧啶碱基 编辑(CBEs) | BE3 | rAPOBEC1 | nCas9D10A | NGG | C4-C8 | C:G>T:A | 实现C>T替换, 无需DSB或提供模板 | Komor et al., | |||
| nCas9-PBE | rAPOBEC1 | nCas9D10A | NGG | C3-C9 | 在植物中实现精准高效的C>T替换 | Zong et al., | |||||
| Target-AID | PmCDA1 | nCas9D10A | NGG | C1-C5 | 具有较好的GC碱基编辑能力 | Nishida et al., | |||||
| dCas9-AIDx | hAID | dCas9 | NCN | C4-C9 | Ma et al., | ||||||
| A3AY130F- CBE_V01 A3AY130F- CBE_V04 | hAPOBEC3AY130F | nCas9D10A | NGG | C4-C15 | 活性窗口拓宽至12 nt | Ren et al., | |||||
| eCDAL | LjCDA1L-4 | nCas9D10A | NGG | C1-C12 | Xu et al., | ||||||
| BE3-PAPAPAP | rAPOBEC1 | nCas9D10A | NGG | C5-C6 | 活性窗口缩小至1-2 nt | Tan et al., | |||||
| PhieCBEs | evorAPOCBEC1 evoFERNY evoCDA1 hA3A | nCas9-NG eCas9n- NG | NG NGG | C-1-C15 | 编辑窗口广 | Zeng et al., | |||||
| BE3R126E BE3R132E YE1-BE3 | rAPOBEC1R126E rAPOBEC1R132E rAPOBEC1W90Y+R126E | nCas9D10A | NGG | C5-C7 | 脱靶率显著降低 | Zuo et al., | |||||
| 腺嘌呤碱基 编辑(ABEs) | ABE7.10 | ecTadA:ecTadA* | nCas9D10A | NGG | A4-A7 | A:T>G:C | 实现A>G替换, 无需DSB或提供模板 | Gaudelli et al., | |||
| PABE | ecTadA:ecTadA* | nCas9D10A | NGG | A4-A8 | 在植物中实现精准高效的A>G替换 | Li et al., | |||||
| ABE-Ps | ecTadA:ecTadA* | nCas9D10A SaCas9 | NGG NNNRRT | A1-A15 | Hua et al., | ||||||
| rBE14 | ecTadA:ecTadA* | nCas9D10A | NGG | A5-A7 | 荧光检测编辑植株 | Yan et al., | |||||
| rABE8e | TadA8eV106W | nCas9D10A nCas9-NG | NG NGG | A5-A6 | 极高的靶点编辑效率和碱基纯合替换效率 | Wei et al., | |||||
| PhieABEs | TadA8e | nCas9-NG SpGn SpRYn | NG NGN NNN | A1-A14 | 编辑效率高, 几近无PAM靶向, 窗口广 | Tan et al., | |||||
| SpRY-ABE8e | ecTadA:ecTadA* | nCas9D10A | NNN | A3-A10 | 几近无PAM靶向 | Ren et al., | |||||
| ABE7.10F148A | ecTadAF148A: ecTadA*F148A | nCas9D10A | NGG | A5-A6 | 活性窗口缩小至1-2 nt | Zhou et al., | |||||
| CG碱基编辑(GBEs) | GBE | AID rAPOBEC1 | nCas9D10A | NGG | C3-C7 | C:G>A:T/G:C | 实现嘧啶与嘌呤间的颠换 | Zhao et al., | |||
| CGBE | rAPOBEC1 | C5-C6 | Chen et al., | ||||||||
| 双碱基编辑(A&CBE) | A&C-BEmax | hAID ecTadA:ecTadA* | nCas9D10A | NGG | C2-C17 A4-A7 | C:G>T:A A:T>G:C | 实现A、C共编辑 | Zhang et al., | |||
| Target-ACE | PmCDA1 ecTadA:ecTadA* | NGG | C1-C10 A4-A8 | Sakata et al., | |||||||
| SPACE | PmCDA1 ecTadA* | NGG | C2-C7 A4-A7 | Grünewald et al., | |||||||
| ACBE | PmCDA1 ecTadA:ecTadA* | NGG | C1-C7 A4-A6 | Xie et al., | |||||||
| STEME | hAPOBEC3A ecTadA:ecTadA* | NG NGD | C1-C17 A4-A8 | 在植物中实现A、 C共编辑 | Li et al., | ||||||
| pDuBE1 | TadA8e | NGG | C1-C10 A2-A9 | Xu et al., | |||||||
| 编辑 技术 | 编辑器 | 脱氨酶/逆转录酶 | Cas蛋白 | PAM (5′-3′) | 活性 窗口 | 编辑类型 | 特点 | 参考文献 | |||
| 先导编辑(PEs) | PE | M-MLV RT | nCas9H840A | NGG | 1-50 | 12种碱基替换 插入(<15 bp) 缺失(<40 bp) | 不受PAM的距离 限制 | Anzalone et al., | |||
| PPE | CaMV RT Retron RT | 在植物中实现PE 应用 | Lin et al., | ||||||||
| pPE2 | M-MLV RT | Xu et al., | |||||||||
| ePPE | M-MLV RT∆RNase H M-MLV RT:NC | 1-92 | 12种碱基替换 插入(<40 bp) 缺失(<100 bp) | 提高了在植物中的 编辑效率 | Zong et al., | ||||||
| 多重 编辑 | SWISSs | rAPOBEC1 ecTadA:ecTadA* | nCas9D10A | NG | C3-C16 A4-A7 | C:G>T:A A:T>G:C Indels | 具有A>G、C>T和Indels三重编辑功能 | Li et al., | |||
| 片段删 除编辑 | AFIDs | hAPOBEC3A hAPOBEC3Bctd | Cas9 | NGG | C1-C14 | 多核苷酸删除 | 精准高效、可预测的多核苷酸缺失 | Wang et al., | |||
表1 各类碱基编辑器的特点
Table 1 Characteristics of all kinds of base editors
| 编辑 技术 | 编辑器 | 脱氨酶/逆转录酶 | Cas蛋白 | PAM (5′-3′) | 活性 窗口 | 编辑类型 | 特点 | 参考文献 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 胞嘧啶碱基 编辑(CBEs) | BE3 | rAPOBEC1 | nCas9D10A | NGG | C4-C8 | C:G>T:A | 实现C>T替换, 无需DSB或提供模板 | Komor et al., | |||
| nCas9-PBE | rAPOBEC1 | nCas9D10A | NGG | C3-C9 | 在植物中实现精准高效的C>T替换 | Zong et al., | |||||
| Target-AID | PmCDA1 | nCas9D10A | NGG | C1-C5 | 具有较好的GC碱基编辑能力 | Nishida et al., | |||||
| dCas9-AIDx | hAID | dCas9 | NCN | C4-C9 | Ma et al., | ||||||
| A3AY130F- CBE_V01 A3AY130F- CBE_V04 | hAPOBEC3AY130F | nCas9D10A | NGG | C4-C15 | 活性窗口拓宽至12 nt | Ren et al., | |||||
| eCDAL | LjCDA1L-4 | nCas9D10A | NGG | C1-C12 | Xu et al., | ||||||
| BE3-PAPAPAP | rAPOBEC1 | nCas9D10A | NGG | C5-C6 | 活性窗口缩小至1-2 nt | Tan et al., | |||||
| PhieCBEs | evorAPOCBEC1 evoFERNY evoCDA1 hA3A | nCas9-NG eCas9n- NG | NG NGG | C-1-C15 | 编辑窗口广 | Zeng et al., | |||||
| BE3R126E BE3R132E YE1-BE3 | rAPOBEC1R126E rAPOBEC1R132E rAPOBEC1W90Y+R126E | nCas9D10A | NGG | C5-C7 | 脱靶率显著降低 | Zuo et al., | |||||
| 腺嘌呤碱基 编辑(ABEs) | ABE7.10 | ecTadA:ecTadA* | nCas9D10A | NGG | A4-A7 | A:T>G:C | 实现A>G替换, 无需DSB或提供模板 | Gaudelli et al., | |||
| PABE | ecTadA:ecTadA* | nCas9D10A | NGG | A4-A8 | 在植物中实现精准高效的A>G替换 | Li et al., | |||||
| ABE-Ps | ecTadA:ecTadA* | nCas9D10A SaCas9 | NGG NNNRRT | A1-A15 | Hua et al., | ||||||
| rBE14 | ecTadA:ecTadA* | nCas9D10A | NGG | A5-A7 | 荧光检测编辑植株 | Yan et al., | |||||
| rABE8e | TadA8eV106W | nCas9D10A nCas9-NG | NG NGG | A5-A6 | 极高的靶点编辑效率和碱基纯合替换效率 | Wei et al., | |||||
| PhieABEs | TadA8e | nCas9-NG SpGn SpRYn | NG NGN NNN | A1-A14 | 编辑效率高, 几近无PAM靶向, 窗口广 | Tan et al., | |||||
| SpRY-ABE8e | ecTadA:ecTadA* | nCas9D10A | NNN | A3-A10 | 几近无PAM靶向 | Ren et al., | |||||
| ABE7.10F148A | ecTadAF148A: ecTadA*F148A | nCas9D10A | NGG | A5-A6 | 活性窗口缩小至1-2 nt | Zhou et al., | |||||
| CG碱基编辑(GBEs) | GBE | AID rAPOBEC1 | nCas9D10A | NGG | C3-C7 | C:G>A:T/G:C | 实现嘧啶与嘌呤间的颠换 | Zhao et al., | |||
| CGBE | rAPOBEC1 | C5-C6 | Chen et al., | ||||||||
| 双碱基编辑(A&CBE) | A&C-BEmax | hAID ecTadA:ecTadA* | nCas9D10A | NGG | C2-C17 A4-A7 | C:G>T:A A:T>G:C | 实现A、C共编辑 | Zhang et al., | |||
| Target-ACE | PmCDA1 ecTadA:ecTadA* | NGG | C1-C10 A4-A8 | Sakata et al., | |||||||
| SPACE | PmCDA1 ecTadA* | NGG | C2-C7 A4-A7 | Grünewald et al., | |||||||
| ACBE | PmCDA1 ecTadA:ecTadA* | NGG | C1-C7 A4-A6 | Xie et al., | |||||||
| STEME | hAPOBEC3A ecTadA:ecTadA* | NG NGD | C1-C17 A4-A8 | 在植物中实现A、 C共编辑 | Li et al., | ||||||
| pDuBE1 | TadA8e | NGG | C1-C10 A2-A9 | Xu et al., | |||||||
| 编辑 技术 | 编辑器 | 脱氨酶/逆转录酶 | Cas蛋白 | PAM (5′-3′) | 活性 窗口 | 编辑类型 | 特点 | 参考文献 | |||
| 先导编辑(PEs) | PE | M-MLV RT | nCas9H840A | NGG | 1-50 | 12种碱基替换 插入(<15 bp) 缺失(<40 bp) | 不受PAM的距离 限制 | Anzalone et al., | |||
| PPE | CaMV RT Retron RT | 在植物中实现PE 应用 | Lin et al., | ||||||||
| pPE2 | M-MLV RT | Xu et al., | |||||||||
| ePPE | M-MLV RT∆RNase H M-MLV RT:NC | 1-92 | 12种碱基替换 插入(<40 bp) 缺失(<100 bp) | 提高了在植物中的 编辑效率 | Zong et al., | ||||||
| 多重 编辑 | SWISSs | rAPOBEC1 ecTadA:ecTadA* | nCas9D10A | NG | C3-C16 A4-A7 | C:G>T:A A:T>G:C Indels | 具有A>G、C>T和Indels三重编辑功能 | Li et al., | |||
| 片段删 除编辑 | AFIDs | hAPOBEC3A hAPOBEC3Bctd | Cas9 | NGG | C1-C14 | 多核苷酸删除 | 精准高效、可预测的多核苷酸缺失 | Wang et al., | |||
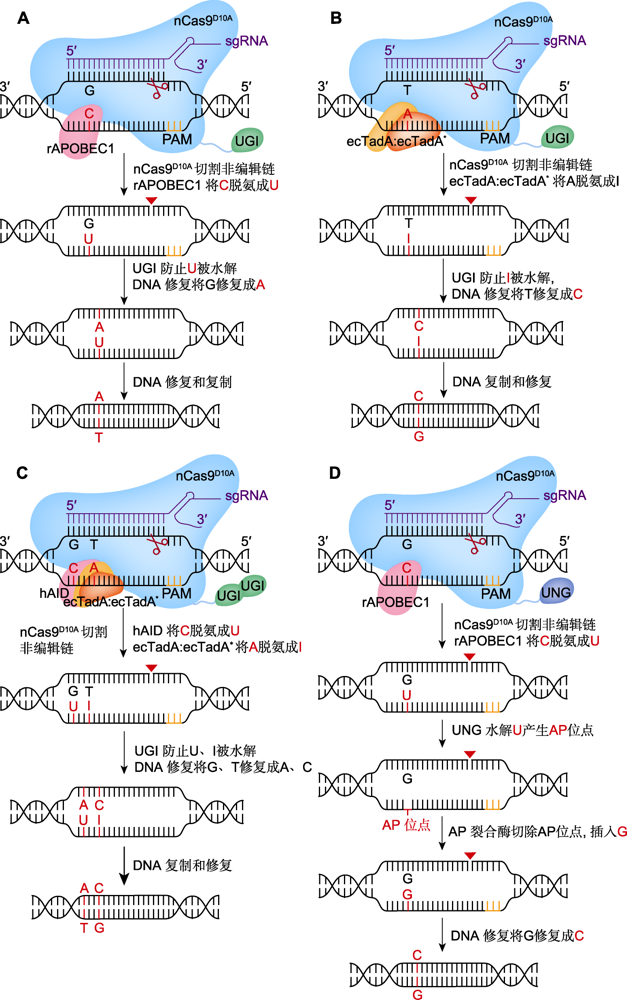
图1 单(CBE、ABE、GBE)、双(A&C-BEmax)碱基编辑器工作原理 (A) BE3介导的C>T替换(在nCas9D10A (蓝色)、胞苷脱氨酶rAPOBEC1 (红色)和尿嘧啶糖基化酶抑制剂(UGI) (绿色)的作用下, 系统BE3将活性窗口中的C脱氨成U, 诱导细胞启动DNA修复实现C到T的替换; 红色三角形表示切口处); (B) ABE7.10介导的A>G替换(在腺苷脱氨酶ecTadA:ecTadA* (黄色和橙色)的作用下, 系统ABE7.10将活性窗口中的A脱氨成I, 并在DNA修复后实现A到G的替换); (C) A&C-BEmax介导的C>T与A>G共替换(在胞嘧啶脱氨酶hAID (红色)、ecTadA:ecTadA*和2个UGI的作用下, 系统A&C-BEmax诱导DNA修复, 实现C到T、A到G的同时替换); (D) GBE介导的C>G颠换(在尿嘧啶-N-糖基化酶(UNG) (深蓝)的作用下, UNG将C脱氨生成的U水解为AP位点, 并在DNA修复后实现C到G的颠换)。PAM: 原间隔序列邻近基序
Figure 1 Technical principles of single (CBE, ABE, GBE) and dual (A&C-BEmax) base editors (A) BE3 mediated C to T base editing (mediated by nCas9D10A (blue), cytidine deaminase rAPOBEC1 (red) and uracil glycosylase inhibitor (UGI) (green), BE3 deaminates C in the active window into U and induces cells to start DNA repair and achieve the replacement of C to T; red triangle represents the notch); (B) ABE7.10 mediated A to G base editing (mediated by adenosine decease ecTadA:ecTadA* (yellow and orange), ABE7.10 deaminates A in the active window into I, and achieves A to G mutation after DNA repair); (C) A&C-BEmax mediated C to T and A to G base editing (mediated by cytidine deaminase hAID (red), ecTadA:ecTadA* and two UGI, A&C-BEmax induces DNA repair and achieves the simultaneous replacement of C to T and A to G); (D) GBE mediated C to G base editing (mediated by uracil-N-glycosylase (UNG) (deep blue), UNG hydrolyzes the U produced by C deamination to AP site, and achieves the transversion from C to G after DNA repair). PAM: Protospacer adjacent motif
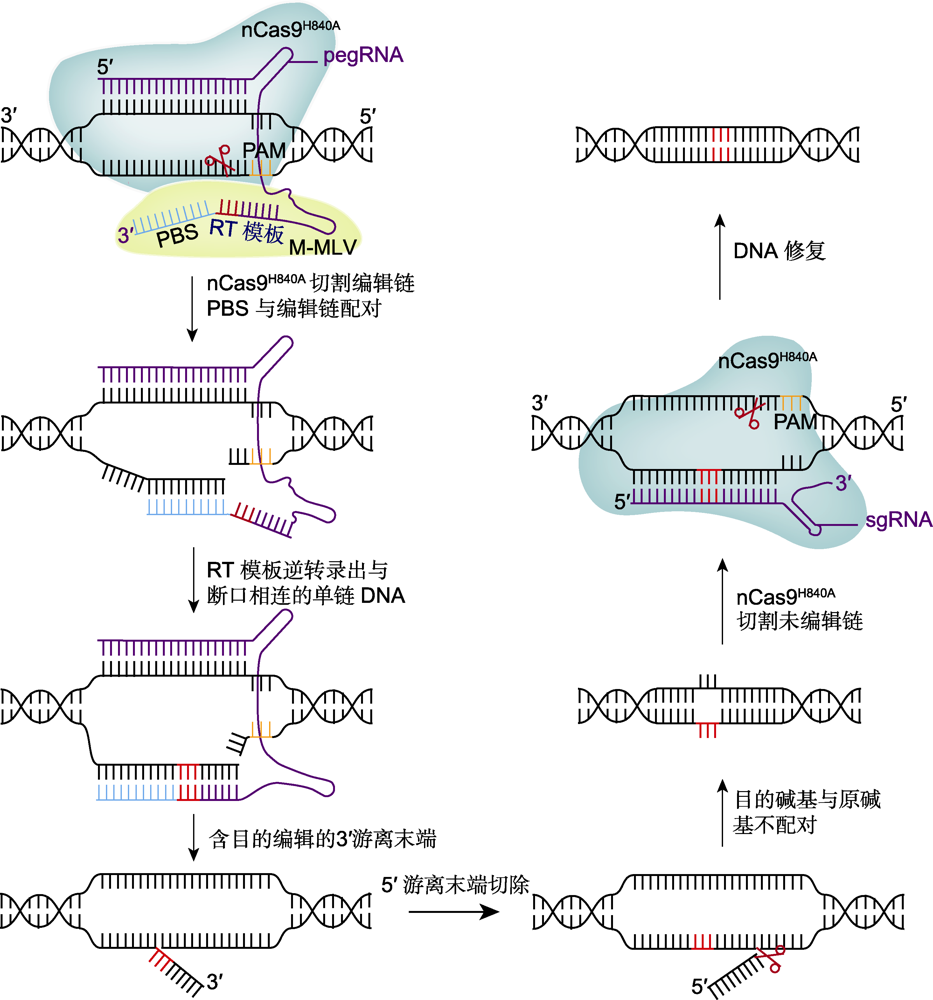
图2 先导编辑器(PE)的工作原理 在nCas9H840A (绿)和逆转录酶M-MLV (黄)的作用下, 系统PE将碱基序列连接到靶位点, 借助DNA修复实现任意碱基的转换以及小片段的插入或删除。PAM: 原间隔序列邻近基序; PBS: 引物结合位点
Figure 2 Technical principles of prime editor (PE) Mediated by nCas9H840A (green) and reverse transcriptase M-MLV (yellow), PE installs the base sequence into the target site, and achieves arbitrary base conversion and precise insertion and deletion of small fragments after DNA repair. PAM: Protospacer adjacent motif; PBS: Primer binding site
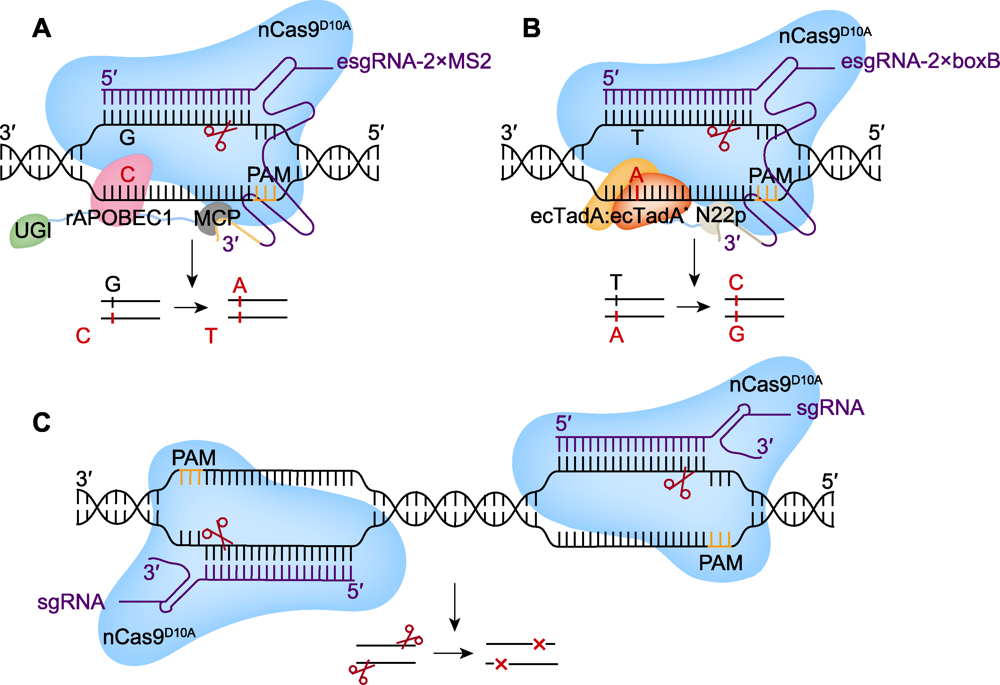
图3 多重编辑系统(SWISS)的工作原理 (A), (B) SWISS系统使用不同的scRNAs (MS2和boxB), 招募融合了相应蛋白(MCP和N22p)的rAPOBEC1或ecTadA:ecTadA*, 实现同时在不同位点的胞嘧啶碱基编辑(CBE)和腺嘌呤碱基编辑(ABE); (C) 使用1对sgRNA在第3个靶点产生DNA双链断裂(DSB), 诱导细胞进行同源定向修复(HDR), 产生随机突变。PAM: 原间隔序列邻近基序; UGI: 尿嘧啶糖基化酶抑制剂
Figure 3 Technical principles of simultaneous and wide-editing induced by a single system (SWISS) (A), (B) Mediated by different scRNAs (MS2 and boxB), SWISS recruits rAPOBEC1 or ecTadA:ecTadA* that combines the corresponding proteins (MCP and N22p) to achieve cytidine (CBE) and adenine (ABE) base editing at different sites; (C) Mediated by a pair of sgRNAs, Cas9 produces double strand break (DSB) at the third target, induces homology-directed repair (HDR) and produces random mutation. PAM: Protospacer adjacent motif; UGI: Uracil glycosylase inhibitor
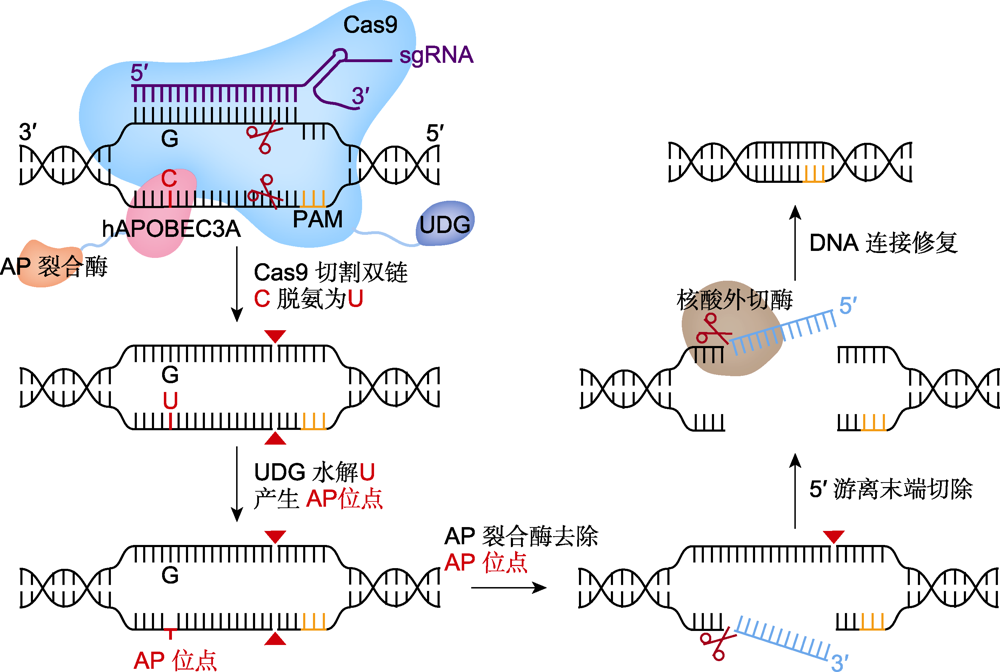
图4 多核苷酸靶向删除系统(AFID)的工作原理 Cas9切割双链产生双链断裂(DSB), 尿嘧啶-DNA-糖基化酶(UDG)将C脱氨生成的U水解为AP位点, 并借助AP裂合酶(橙红色)和核酸外切酶(棕色), 实现靶点C到DSB切口之间的多核苷酸删除。PAM: 原间隔序列邻近基序
Figure 4 Technical principles of APOBEC-Cas9 fusion-induced deletion systems (AFIDs) Cas9 cuts both strands to form adouble strand break (DSB), uracil-DNA-glycosylase (UDG) hydrolyzes the U produced by C deamination to AP site, and achieve the polynucleotide deletion between target C and DSB with the help of AP lyase (orange red) and exonuclease (brown). PAM: Protospacer adjacent motif
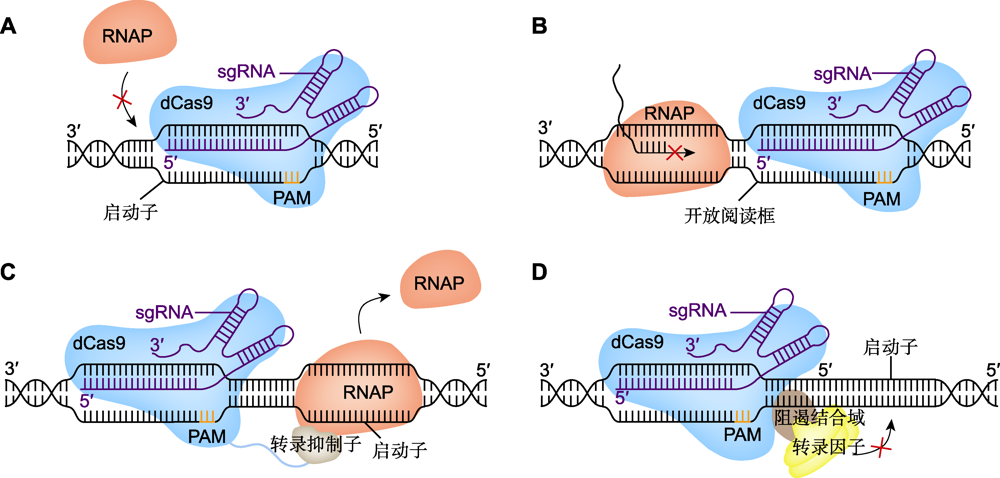
图5 CRISPR干扰系统的工作原理 (A) dCas9阻止RNA聚合酶(RNAP)结合基因启动子, sgRNA介导dCas9结合目的基因启动子, 使RNAP (红色)无法与该基因启动子结合并进行转录; (B) dCas9阻断RNAP的转录延伸, sgRNA介导dCas9结合目的基因开放阅读框(ORF), 使RNAP无法继续转录延伸; (C) 转录抑制子阻止RNAP结合基因启动子, dCas9与转录抑制子(灰色)融合, 抑制子会阻止RNAP与目的基因启动子的结合; (D) 阻遏结合域(RBD)阻止目的基因的转录激活, dCas9与RBD (棕色)融合, RBD阻断转录因子(TFs)与目的基因的结合, 并与TFs结合形成强阻遏物抑制基因的转录表达。PAM: 原间隔序列邻近基序
Figure 5 Technical principles of CRISPR interference system (A) dCas9 prevents RNA polymerase (RNAP) from binding to gene promoter, sgRNA mediated dCas9 binding to the target gene promoter, so that RNAP (red) can not bind to the gene promoter to start transcribing; (B) dCas9 blocks the transcriptional extension of RNAP, sgRNA mediates dCas9 binding to the target gene open reading frame (ORF), making RNAP unable to continue transcriptional extension; (C) Transcriptional repressors prevent RNAP from binding to gene promoters, dCas9 fuses with the transcription inhibitor (gray), which prevents the binding of RNAP to the promoter of the target gene; (D) The repressor binding domain (RBD) blocks the transcriptional activation of the target gene, dCas9 fuses with the RBD (brown), RBD blocked the binding of transcription factors (TFs) to the target gene and combined with TFs to form a strong repressor to inhibit the transcription of the gene. PAM: Protospacer adjacent motif
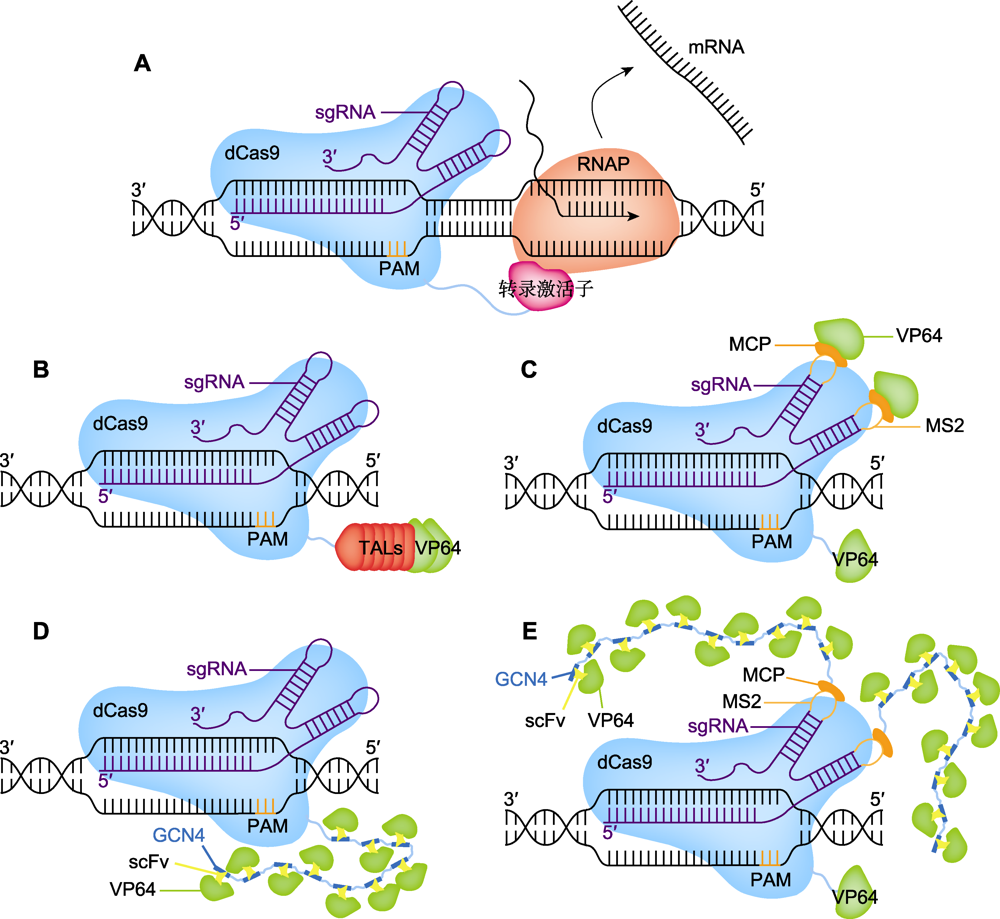
图6 CRISPR激活系统的工作原理 (A) 转录激活子激活目的基因转录, dCas9与转录激活子(洋红色)融合, 招募RNA聚合酶(RNAP)并激活目的基因的转录; (B)-(E) 不同CRISPRa系统示意图, 包括TALs (橙红色)、VP64 (绿色)、MS2 (浅黄色)、MCP (深黄色)、GCN4 (湛蓝色)和scFv (明黄色)。PAM: 原间隔序列邻近基序
Figure 6 Technical principles of CRISPR activation system (A) Transcriptional activator activates the transcription of the target gene, dCas9 fuses with transcription activator (magenta) to recruit RNA polymerase (RNAP) and activates the transcription of the target gene; (B)-(E) Schematic diagram of different CRISPRa systems, including TALs (orange red), VP64 (green), MS2 (light yellow), MCP (deep yellow), GCN4 (azure blue) and scFv (bright yellow). PAM: Protospacer adjacent motif

图7 基因组修饰系统去甲基化和甲基化的工作原理 (A) DNA去甲基化修饰, dCas9-SunTag系统招募人脱甲基酶TET1cd (棕色), 在目的基因的启动子区使DNA去甲基化, Me表示甲基化; (B) DNA甲基化修饰, dCas9-SunTag系统招募烟草DRM甲基转移酶催化结构域NtDRMcd (绿色), 在目的基因启动子区使DNA甲基化。PAM: 原间隔序列邻近基序
Figure 7 Technical principles of demethylation and methylation of genome modification systems (A) DNA demethylation modification, dCas9-SunTag system recruits human demethylase TET1cd (brown) to demethylate DNA in the promoter region of the target gene, Me represents methylation; (B) DNA methylation modification, dCas9-SunTag system recruits tobacco DRM methyltransferase catalytic domain NtDRMcd (green) to methylate DNA in the promoter region of the target gene. PAM: Protospacer adjacent motif
| 物种 | 品种 | 靶基因 | 转化方法 | 编辑 技术 | 脱氨酶/逆转录酶 | 编辑类型 | 编辑 窗口 | 编辑效率(%) | 参考文献 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 水稻(Oryza sativa) | 中花11 | CDC48 NRT1.1B-T1 | 农杆菌AGL1 | CBEs | hAPOBEC3A | C:G>T:A | C2-C16 | 44.1- 82.9 | Zong et al., | |||
| 日本晴 | CDC48 | 农杆菌AGL1 | rAPOBEC1 | C:G>T:A | C3-C8 | 43.48 | Zong et al., | |||||
| Kitaake | Pi-d2 FLS2 | 农杆菌EHA105 | hAID*Δ | C:G>T:A | C3-C7 | 30.8-57 | Ren et al., | |||||
| 中花11 | ACC-T1 ALS-T1 CDC48-T3 DEP1-T1/2 NRT1.1B-T1 | 农杆菌AGL1 | ABEs | ecTadA: ecTadA* | A:T>G:C | A4-A8 | 15.8- 59.1 | Li et al., | ||||
| 日本晴 | SPL14/16/17/18 SLR1 | 农杆菌EHA105 | ecTadA: ecTadA* | A:T>G:C | A5-A14 | 12.5- 61.3 | Hua et al., | |||||
| Kitaake | SERK2 WRKY45 | 农杆菌EHA105 | ecTadA: ecTadA* | A:T>G:C | A4-A7 | 32.05- 62.26 | Yan et al., | |||||
| 中花11 | ACC | 农杆菌AGL1 | STEMEs | hAPOBEC3A ecTadA: ecTadA* | C:G>T:A; A:T>G:C | C1-C17 A4-A8 | 3.84 | Li et al., | ||||
| 中花11 | CDC48-T1 ALS-T2 | 农杆菌EHA105 | PEs | M-MLV | G:C>T:A 插入(≤3 bp) 删除(≤6 bp) | N1-N6 | 2.6-21.8 | Lin et al., | ||||
| 日本晴 | ALS-1/2 ACC-1 DEP1 | 农杆菌EHA105 | PEs | M-MLV | C:G>T:A A:T>G:C G:C>T:A G:C>C:G G:C>A:T T:A>A:T A:T>C:G | N18-N33 | 1.7-26 | Xu et al., | ||||
| 物种 | 品种 | 靶基因 | 转化方法 | 编辑 技术 | 脱氨酶/逆转录酶 | 编辑类型 | 编辑 窗口 | 编辑效率(%) | 参考文献 | |||
| 中花11 | ALS-T2 ACC-T2 BADH-indels | 农杆菌AGL1 | SWISS | hAPOBEC3A ecTadA: ecTadA* | C:G>T:A A:T>G:C C:G>G:C 删除(≤45 bp) | C6-C7 A4-A7 | 7.3 | Li et al., | ||||
| 中花11 | CDC48-T2 SPL14 SWEET14 | 农杆菌AGL1 | AFID | hAPOBEC3A | 删除(≤16 bp) | C2-C17 | 22.2- 55.8 | Wang et al., | ||||
| 小麦(Triticum aestivum) | Kenong 199 | ALS MTL | 基因枪 | CBEs | hAPOBEC3A | C:G>T:A | C-9-C13 | 16.7- 22.5 | Zong et al., | |||
| Bobwhite | LOX2-S1 | 基因枪 | CBEs | rAPOBEC1 | C:G>T:A | C3-C9 | 1.25 | Zong et al., | ||||
| Kenong 199 | ALS | 基因枪 | CBEs | rAPOBEC1 | C:G>T:A | C-1-C7 | 22-78 | Zhang et al., | ||||
| Kenong 199 | DEP1 GW2 | 基因枪 | ABEs | ecTadA: ecTadA* | A:T>G:C | A5-A8 | 0.4-1.1 | Li et al., | ||||
| Kenong 199 | miR396 GASR6 | 基因枪 | AFID | hAPOBEC3A | 删除(≤35 bp) | C-12-C23 | 25-37.5 | Wang et al., | ||||
| 番茄(Solanum lycopersicum) | WVA106 | ALS1 | 农杆菌C58 pGV2260 | CBEs | PmCDA1 | C:G>T:A | C7 | 34.7 | Veillet et al., | |||
| WVA106 | ALS1 | 农杆菌C58 pGV2260 | CBEs | PmCDA1 | C:G>T:A | C1-C8 | 20.59 | Veillet et al., | ||||
| Micro- Tom | DELLA | 农杆菌 | CBEs | PmCDA1 | C:G>T:A | C1-C3 | 50.5 | Shimatani et al., | ||||
| 马铃薯 (S. tuberosum) | Désirée | GBSS1 DMR6-1 | PEG | CBEs | hAPOBEC3A | C:G>T:A | C3-C10 | 8-16.67 | Veillet et al., | |||
| Désirée | GBSS-T6 | PEG | CBEs | hAPOBEC3A | C:G>T:A | C1-C13 | 6.5 | Jiang et al., | ||||
| Désirée | ALS1 | 农杆菌C58 pGV2260 | CBEs | PmCDA1 | C:G>T:A | C1-C8 | 25 | Veillet et al., | ||||
| 大豆 (Glycine max) | Jack | FT2a FT4 | 农杆菌 | CBEs | rAPOBEC1 | C:G>T:A | C6-C7 | 5.88- 18.2 | Cai et al., | |||
| 棉花(Gossypium hirsutum) | Jin668 | CLA PEBP | 农杆菌GV3101 | CBEs | rAPOBEC1 | C:G>T:A | C4-C8 | 26.67- 57.78 | Qin et al., | |||
| 玉米 (Zea mays) | Zong31 | CENH3 | 农杆菌AGL1 | CBEs | rAPOBEC1 | C:G>T:A | C3-C8 | 10.1 | Zong et al., | |||
| ND73 | ALS1/2 | 农杆菌LBA4404/ pVS1-VIR2 | PEs | CmYLCV | C:G>T:A A:T>C:G T:A>G:C T:A>C:G C:G>A:T G:C>T:A G:C>A:T G:C>C:G | N3-N46 | 6.5-53.2 | Jiang et al., | ||||
| 油菜(Brassica napus) | J9712 | ALS1 | 农杆菌 | CBEs | rAPOBEC1 | C:G>T:A | C5-C7 | 1.8 | Wu et al., | |||
| 西瓜(Citrullus lanatus) | ZG94 | ALS | 农杆菌EHA105 | CBEs | rAPOBEC1 | C:G>T:A | C7-C8 | 23 | Tian et al., | |||
表2 碱基编辑器在作物中的应用
Table 2 Application of base editors in crop
| 物种 | 品种 | 靶基因 | 转化方法 | 编辑 技术 | 脱氨酶/逆转录酶 | 编辑类型 | 编辑 窗口 | 编辑效率(%) | 参考文献 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 水稻(Oryza sativa) | 中花11 | CDC48 NRT1.1B-T1 | 农杆菌AGL1 | CBEs | hAPOBEC3A | C:G>T:A | C2-C16 | 44.1- 82.9 | Zong et al., | |||
| 日本晴 | CDC48 | 农杆菌AGL1 | rAPOBEC1 | C:G>T:A | C3-C8 | 43.48 | Zong et al., | |||||
| Kitaake | Pi-d2 FLS2 | 农杆菌EHA105 | hAID*Δ | C:G>T:A | C3-C7 | 30.8-57 | Ren et al., | |||||
| 中花11 | ACC-T1 ALS-T1 CDC48-T3 DEP1-T1/2 NRT1.1B-T1 | 农杆菌AGL1 | ABEs | ecTadA: ecTadA* | A:T>G:C | A4-A8 | 15.8- 59.1 | Li et al., | ||||
| 日本晴 | SPL14/16/17/18 SLR1 | 农杆菌EHA105 | ecTadA: ecTadA* | A:T>G:C | A5-A14 | 12.5- 61.3 | Hua et al., | |||||
| Kitaake | SERK2 WRKY45 | 农杆菌EHA105 | ecTadA: ecTadA* | A:T>G:C | A4-A7 | 32.05- 62.26 | Yan et al., | |||||
| 中花11 | ACC | 农杆菌AGL1 | STEMEs | hAPOBEC3A ecTadA: ecTadA* | C:G>T:A; A:T>G:C | C1-C17 A4-A8 | 3.84 | Li et al., | ||||
| 中花11 | CDC48-T1 ALS-T2 | 农杆菌EHA105 | PEs | M-MLV | G:C>T:A 插入(≤3 bp) 删除(≤6 bp) | N1-N6 | 2.6-21.8 | Lin et al., | ||||
| 日本晴 | ALS-1/2 ACC-1 DEP1 | 农杆菌EHA105 | PEs | M-MLV | C:G>T:A A:T>G:C G:C>T:A G:C>C:G G:C>A:T T:A>A:T A:T>C:G | N18-N33 | 1.7-26 | Xu et al., | ||||
| 物种 | 品种 | 靶基因 | 转化方法 | 编辑 技术 | 脱氨酶/逆转录酶 | 编辑类型 | 编辑 窗口 | 编辑效率(%) | 参考文献 | |||
| 中花11 | ALS-T2 ACC-T2 BADH-indels | 农杆菌AGL1 | SWISS | hAPOBEC3A ecTadA: ecTadA* | C:G>T:A A:T>G:C C:G>G:C 删除(≤45 bp) | C6-C7 A4-A7 | 7.3 | Li et al., | ||||
| 中花11 | CDC48-T2 SPL14 SWEET14 | 农杆菌AGL1 | AFID | hAPOBEC3A | 删除(≤16 bp) | C2-C17 | 22.2- 55.8 | Wang et al., | ||||
| 小麦(Triticum aestivum) | Kenong 199 | ALS MTL | 基因枪 | CBEs | hAPOBEC3A | C:G>T:A | C-9-C13 | 16.7- 22.5 | Zong et al., | |||
| Bobwhite | LOX2-S1 | 基因枪 | CBEs | rAPOBEC1 | C:G>T:A | C3-C9 | 1.25 | Zong et al., | ||||
| Kenong 199 | ALS | 基因枪 | CBEs | rAPOBEC1 | C:G>T:A | C-1-C7 | 22-78 | Zhang et al., | ||||
| Kenong 199 | DEP1 GW2 | 基因枪 | ABEs | ecTadA: ecTadA* | A:T>G:C | A5-A8 | 0.4-1.1 | Li et al., | ||||
| Kenong 199 | miR396 GASR6 | 基因枪 | AFID | hAPOBEC3A | 删除(≤35 bp) | C-12-C23 | 25-37.5 | Wang et al., | ||||
| 番茄(Solanum lycopersicum) | WVA106 | ALS1 | 农杆菌C58 pGV2260 | CBEs | PmCDA1 | C:G>T:A | C7 | 34.7 | Veillet et al., | |||
| WVA106 | ALS1 | 农杆菌C58 pGV2260 | CBEs | PmCDA1 | C:G>T:A | C1-C8 | 20.59 | Veillet et al., | ||||
| Micro- Tom | DELLA | 农杆菌 | CBEs | PmCDA1 | C:G>T:A | C1-C3 | 50.5 | Shimatani et al., | ||||
| 马铃薯 (S. tuberosum) | Désirée | GBSS1 DMR6-1 | PEG | CBEs | hAPOBEC3A | C:G>T:A | C3-C10 | 8-16.67 | Veillet et al., | |||
| Désirée | GBSS-T6 | PEG | CBEs | hAPOBEC3A | C:G>T:A | C1-C13 | 6.5 | Jiang et al., | ||||
| Désirée | ALS1 | 农杆菌C58 pGV2260 | CBEs | PmCDA1 | C:G>T:A | C1-C8 | 25 | Veillet et al., | ||||
| 大豆 (Glycine max) | Jack | FT2a FT4 | 农杆菌 | CBEs | rAPOBEC1 | C:G>T:A | C6-C7 | 5.88- 18.2 | Cai et al., | |||
| 棉花(Gossypium hirsutum) | Jin668 | CLA PEBP | 农杆菌GV3101 | CBEs | rAPOBEC1 | C:G>T:A | C4-C8 | 26.67- 57.78 | Qin et al., | |||
| 玉米 (Zea mays) | Zong31 | CENH3 | 农杆菌AGL1 | CBEs | rAPOBEC1 | C:G>T:A | C3-C8 | 10.1 | Zong et al., | |||
| ND73 | ALS1/2 | 农杆菌LBA4404/ pVS1-VIR2 | PEs | CmYLCV | C:G>T:A A:T>C:G T:A>G:C T:A>C:G C:G>A:T G:C>T:A G:C>A:T G:C>C:G | N3-N46 | 6.5-53.2 | Jiang et al., | ||||
| 油菜(Brassica napus) | J9712 | ALS1 | 农杆菌 | CBEs | rAPOBEC1 | C:G>T:A | C5-C7 | 1.8 | Wu et al., | |||
| 西瓜(Citrullus lanatus) | ZG94 | ALS | 农杆菌EHA105 | CBEs | rAPOBEC1 | C:G>T:A | C7-C8 | 23 | Tian et al., | |||
| [1] |
苏钺凯, 邱镜仁, 张晗, 宋振巧, 王建华 (2019). CRISPR/ Cas9系统在植物基因组编辑中技术改进与创新的研究进展. 植物学报 54, 385-395.
DOI |
| [2] | 谭禄宾, 孙传清 (2021). 四倍体野生稻快速驯化: 启动人类新农业文明. 植物学报 56, 134-137. |
| [3] |
王影, 李相敢, 邱丽娟 (2018). CRISPR/Cas9基因组定点编辑中脱靶现象的研究进展. 植物学报 53, 528-541.
DOI |
| [4] |
谢先荣, 曾栋昌, 谭健韬, 祝钦泷, 刘耀光 (2021). 基于CRISPR编辑系统的DNA片段删除技术. 植物学报 56, 44-49.
DOI |
| [5] |
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149-157.
DOI URL |
| [6] |
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709-1712.
DOI PMID |
| [7] |
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551-2561.
DOI URL |
| [8] |
Bolukbasi MF, Gupta A, Wolfe SA (2016). Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat Methods 13, 41-50.
DOI PMID |
| [9] |
Cai WQ, Wang M (2019). Engineering nucleic acid chemistry for precise and controllable CRISPR/Cas9 genome editing. Sci Bull 64, 1841-1849.
DOI URL |
| [10] |
Cai YP, Chen L, Zhang Y, Yuan S, Su Q, Sun S, Wu CX, Yao WW, Han TF, Hou WS (2020). Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol J 18, 1996-1998.
DOI URL |
| [11] |
Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA (2017). Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407-410.
DOI URL |
| [12] |
Chen LW, Park JE, Paa P, Rajakumar PD, Prekop HT, Chew YT, Manivannan SN, Chew WL (2021). Programmable C:G to G:C genome editing with CRISPR- Cas9-directed base excision repair proteins. Nat Commun 12, 1384.
DOI URL |
| [13] |
Chow RD, Chen JS, Shen J, Chen SD (2021). A web tool for the design of prime-editing guide RNAs. Nat Biomed Eng 5, 190-194.
DOI URL |
| [14] |
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757-761.
DOI PMID |
| [15] |
Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190, 1390-1400.
DOI URL |
| [16] |
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE (2016). Optimized sgRNA design to maximize activity and minimize off- target effects of CRISPR-Cas9. Nat Biotechnol 34, 184-191.
DOI PMID |
| [17] |
Duan JZ, Lu GQ, Xie Z, Lou ML, Luo J, Guo L, Zhang Y (2014). Genome-wide identification of CRISPR/Cas9 off- targets in human genome. Cell Res 24, 1009-1012.
DOI URL |
| [18] |
Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lécrivain AL, Bzdrenga J, Koonin EV, Charpentier E (2014). Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res 42, 2577-2590.
DOI PMID |
| [19] | Gallego-Bartolomé J, Gardiner J, Liu WL, Papikian A, Ghoshal B, Kuo HY, Zhao JMC, Segal DJ, Jacobsen SE (2018). Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc Natl Acad Sci USA 115, E2125-E2134. |
| [20] | Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109, E2579-E2586. |
| [21] |
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017). Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature 551, 464-471.
DOI URL |
| [22] |
Grünewald J, Zhou RH, Lareau CA, Garcia SP, Iyer S, Miller BR, Langner LM, Hsu JY, Aryee MJ, Joung JK (2020). A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat Biotechnol 38, 861-864.
DOI PMID |
| [23] | Hess GT, Frésard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, Bassik MC (2016). Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods 13, 1036-1042. |
| [24] |
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li YQ, Fine EJ, Wu XB, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827-832.
DOI URL |
| [25] |
Hu B, Wang W, Ou SJ, Tang JY, Li H, Che RH, Zhang ZH, Chai XY, Wang HR, Wang YQ, Liang CZ, Liu LC, Piao ZZ, Deng QY, Deng K, Xu C, Liang Y, Zhang LH, Li LG, Chu CC (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47, 834-838.
DOI |
| [26] |
Hua K, Tao XP, Yuan FT, Wang D, Zhu JK (2018). Precise A·T to G·C base editing in the rice genome. Mol Plant 11, 627-630.
DOI URL |
| [27] |
Huang XZ, Qian Q, Liu ZB, Sun HY, He SY, Luo D, Xia GM, Chu CC, Li JY, Fu XD (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41, 494-497.
DOI URL |
| [28] |
Ines F, Anaïs LR, Krzysztof C, Kira SM, Anne LL, Janek B, Eugene VK, Emmanuelle C (2014). Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res 42, 2577-2590.
DOI URL |
| [29] |
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169, 5429-5433.
DOI PMID |
| [30] |
Jansen R, Gaastra W, Schouls LM (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43, 1565-1575.
PMID |
| [31] |
Jiang TT, Zhang XO, Weng ZP, Xue W (2022). Deletion and replacement of long genomic sequences using prime editing. Nat Biotechnol 40, 227-234.
DOI URL |
| [32] |
Jiang WY, Bikard D, Cox D, Zhang F, Marraffini LA (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31, 233-239.
DOI URL |
| [33] |
Jiang YY, Chai YP, Lu MH, Han XL, Lin QP, Zhang Y, Zhang Q, Zhou Y, Wang XC, Gao CX, Chen QJ (2020). Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol 21, 257.
DOI URL |
| [34] |
Jiao YQ, Wang YH, Xue DW, Wang J, Yan MX, Liu GF, Dong GJ, Zeng DL, Lu ZF, Zhu XD, Qian Q, Li JY (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42, 541-544.
DOI URL |
| [35] |
Jin S, Fei HY, Zhu ZX, Luo YF, Liu JX, Gao SH, Zhang F, Chen YH, Wang YP, Gao CX (2020). Rationally designed APOBEC3B Cytosine base editors with improved specificity. Mol Cell 79, 728-740.
DOI URL |
| [36] |
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821.
DOI URL |
| [37] |
Kim YG, Cha J, Chandrasegaran S (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA 93, 1156-1160.
DOI URL |
| [38] |
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng ZL, Joung JK (2016). High-fidelity CRISPR- Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490-495.
DOI URL |
| [39] |
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420-424.
DOI URL |
| [40] |
Kuang YJ, Li SF, Ren B, Yan F, Spetz C, Li XJ, Zhou XP, Zhou HB (2020). Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol Plant 13, 565-572.
DOI URL |
| [41] |
Li C, Zhang R, Meng XB, Chen S, Zong Y, Lu CJ, Qiu JL, Chen YH, Li JY, Gao CX (2020a). Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat Biotechnol 38, 875-882.
DOI URL |
| [42] |
Li C, Zong Y, Jin S, Zhu HC, Lin DX, Li SN, Qiu JL, Wang YP, Gao CX (2020b). SWISS: multiplexed orthogonal genome editing in plants with a Cas9 nickase and engineered CRISPR RNA scaffolds. Genome Biol 21, 141.
DOI URL |
| [43] | Li C, Zong Y, Wang YP, Jin S, Zhang DB, Song QN, Zhang R, Gao CX (2018a). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19, 59. |
| [44] |
Li SN, Lin DX, Zhang YW, Deng M, Chen YX, Lv B, Li BS, Lei Y, Wang YP, Zhao L, Liang YT, Liu JX, Chen KL, Liu ZY, Xiao J, Qiu JL, Gao CX (2022). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455-460.
DOI URL |
| [45] |
Li TD, Yang XP, Yu Y, Si XM, Zhai XW, Zhang HW, Dong WX, Gao CX, Xu C (2018b). Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol 36, 1160-1163.
DOI URL |
| [46] |
Li ZX, Zhang DD, Xiong XY, Yan BY, Xie W, Sheen J, Li JF (2017). A potent Cas9-derived gene activator for plant and mammalian cells. Nat Plants 3, 930-936.
DOI URL |
| [47] |
Liang Z, Chen KL, Li TD, Zhang Y, Wang YP, Zhao Q, Liu JX, Zhang HW, Liu CM, Ran YD, Gao CX (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8, 14261.
DOI PMID |
| [48] |
Lin QP, Zong Y, Xue CX, Wang SX, Jin S, Zhu ZX, Wang YP, Anzalone AV, Raguram A, Doman JL, Liu DR, Gao CX (2020). Prime genome editing in rice and wheat. Nat Biotechnol 38, 582-585.
DOI URL |
| [49] |
Liu H, Ding YD, Zhou YQ, Jin WQ, Xie KB, Chen LL (2017). CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10, 530-532.
DOI URL |
| [50] |
Liu MZ, Zhang WW, Xin CC, Yin JH, Shang YF, Ai C, Li JX, Meng FL, Hu JZ (2021). Global detection of DNA repair outcomes induced by CRISPR-Cas9. Nucleic Acids Res 49, 8732-8742.
DOI URL |
| [51] |
Lowder LG, Zhang DW, Baltes NJ, Paul JW, Tang X, Zheng XL, Voytas DF, Hsieh TF, Zhang Y, Qi YP (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169, 971-985.
DOI PMID |
| [52] |
Lowder LG, Zhou JP, Zhang YX, Malzahn A, Zhong ZH, Hsieh TF, Voytas DF, Zhang Y, Qi YP (2018). Robust transcriptional activation in plants using multiplexed CRISPR- Act2.0 and mTALE-Act systems. Mol Plant 11, 245-256.
DOI PMID |
| [53] |
Lu Y, Wang JY, Chen B, Mo SD, Lian L, Luo YM, Ding DH, Ding YH, Cao Q, Li YC, Li Y, Liu GZ, Hou QQ, Cheng TT, Wei JT, Zhang YR, Chen GW, Song C, Hu Q, Sun S, Fan GY, Wang YT, Liu ZT, Song BA, Zhu JK, Li HR, Jiang LJ (2021). A donor-DNA-free CRISPR/Cas-based approach to gene knock-up in rice. Nat Plants 7, 1445-1452.
DOI URL |
| [54] |
Ma XN, Zhang XY, Liu HM, Li ZH (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat Plants 6, 773-779.
DOI URL |
| [55] |
Ma YQ, Zhang JY, Yin WJ, Zhang ZC, Song Y, Chang X (2016). Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 13, 1029-1035.
DOI |
| [56] |
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13, 722-736.
DOI PMID |
| [57] |
Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo J, Norville JE, Church GM (2013). RNA-guided human genome engineering via Cas9. Science 339, 823-826.
DOI URL |
| [58] |
Mok BY, de Moraes MH, Zeng J, Bosch DE, Kotrys AV, Raguram A, Hsu F, Radey MC, Peterson SB, Mootha VK, Mougous JD, Liu DR (2020). A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631-637.
DOI URL |
| [59] |
Molla KA, Sretenovic S, Bansal KC, Qi YP (2021). Precise plant genome editing using base editors and prime editors. Nat Plants 7, 1166-1187.
DOI URL |
| [60] |
Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV, An MR, Newby GA, Chen JC, Hsu A, Liu DR (2022). Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol 40, 402-410.
DOI URL |
| [61] | Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729. |
| [62] |
Nishimasu H, Shi X, Ishiguro S, Gao LY, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O (2018). Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259-1262.
DOI PMID |
| [63] |
Oakes BL, Fellmann C, Rishi H, Taylor KL, Ren SM, Nadler DC, Yokoo R, Arkin AP, Doudna JA, Savage DF (2019). CRISPR-Cas9 circular permutants as programmable scaffolds for genome modification. Cell 176, 254-267.
DOI URL |
| [64] |
Oliva R, Ji CH, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T, Eom JS, Li CH, Nguyen H, Liu B, Auguy F, Sciallano C, Luu VT, Dossa GS, Cunnac S, Schmidt SM, Slamet-Loedin IH, Cruz CV, Szurek B, Frommer WB, White FF, Yang B (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37, 1344-1350.
DOI URL |
| [65] |
Pan CT, Wu XC, Markel K, Malzahn AA, Kundagrami N, Sretenovic S, Zhang YX, Cheng YH, Shih PM, Qi YP (2021). CRISPR-Act3.0 for highly efficient multiplexed gene activation in plants. Nat Plants 7, 942-953.
DOI URL |
| [66] |
Papikian A, Liu WL, Gallego-Bartolomé J, Jacobsen SE (2019). Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun 10, 729.
DOI PMID |
| [67] | Pattanayak V, Lin S, Guilinger JP, Ma EB, Doudna JA, Liu DR (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31, 839-843. |
| [68] |
Piatek A, Ali Z, Baazim H, Li LX, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM (2015). RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13, 578-589.
DOI URL |
| [69] |
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173-1183.
DOI URL |
| [70] |
Qin L, Li JY, Wang QQ, Xu ZP, Sun L, Alariqi M, Manghwar H, Wang GY, Li B, Ding X, Rui HP, Huang HM, Lu TL, Lindsey K, Daniell H, Zhang XL, Jin SX (2020). High-efficient and precise base editing of C·G to T·A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol J 18, 45-56.
DOI URL |
| [71] |
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu XB, Makarova KS, Koonin EV, Sharp PA, Zhang F (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186-191.
DOI URL |
| [72] |
Ren B, Yan F, Kuang YJ, Li N, Zhang DW, Zhou XP, Lin HH, Zhou HB (2018). Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9- guided hyperactive hAID mutant. Mol Plant 11, 623-626.
DOI URL |
| [73] |
Ren QR, Sretenovic S, Liu GQ, Zhong ZH, Wang JH, Huang L, Tang X, Guo YC, Liu L, Wu YC, Zhou J, Zhao YX, Yang H, He Y, Liu SS, Yin DS, Mayorga R, Zheng XL, Zhang T, Qi YP, Zhang Y (2021a). Improved plant cytosine base editors with high editing activity, purity, and specificity. Plant Biotechnol J 19, 2052-2068.
DOI URL |
| [74] |
Ren QR, Sretenovic S, Liu SS, Tang X, Huang L, He Y, Liu L, Guo YC, Zhong ZH, Liu GQ, Cheng YH, Zheng XL, Pan CT, Yin DS, Zhang YX, Li WF, Qi LW, Li CH, Qi YP, Zhang Y (2021b). PAM-less plant genome editing using a CRISPR-SpRY toolbox. Nat Plants 7, 25-33.
DOI URL |
| [75] |
Sakata RC, Ishiguro S, Mori H, Tanaka M, Seki M, Masuyama N, Nishida K, Nishimasu H, Kondo A, Nureki O, Tomita M, Aburatani H, Yachie N (2019). A single CRISPR base editor to induce simultaneous C-to-T and A-to-G mutations. BioRxiv doi: 10.1101/729269.
DOI |
| [76] |
Sandhya D, Jogam P, Allini VR, Abbagani S, Alok A (2020). The present and potential future methods for delivering CRISPR/Cas9 components in plants. J Genet Eng Biotechnol 18, 25.
DOI PMID |
| [77] |
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35, 441-443.
DOI PMID |
| [78] |
Slaymaker IM, Gao LY, Zetsche B, Scott DA, Yan WX, Zhang F (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84-88.
DOI PMID |
| [79] |
Sun HY, Qian Q, Wu K, Luo JJ, Wang SS, Zhang CW, Ma YF, Liu Q, Huang XZ, Yuan QB, Han RX, Zhao M, Dong GJ, Guo LB, Zhu XD, Gou ZH, Wang W, Wu YJ, Lin HX, Fu XD (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet 46, 652-656.
DOI URL |
| [80] |
Symington LS, Gautier J (2011). Double-strand break end resection and repair pathway choice. Annu Rev Genet 45, 247-271.
DOI PMID |
| [81] |
Tan JJ, Zhang F, Karcher D, Bock R (2019). Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat Commun 10, 439.
DOI URL |
| [82] |
Tan JT, Zeng DC, Zhao YC, Wang YX, Liu TL, Li SC, Xue Y, Luo YY, Xie XR, Chen LT, Liu YG, Zhu QL (2022). PhieABEs: a PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol J 20, 934-943.
DOI URL |
| [83] |
Thuronyi BW, Koblan LW, Levy JM, Yeh WH, Zheng C, Newby GA, Wilson C, Bhaumik M, Shubina-Oleinik O, Holt JR, Liu DR (2019). Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol 37, 1070-1079.
DOI URL |
| [84] |
Tian SW, Jiang LJ, Cui XX, Zhang J, Guo SG, Li MY, Zhang HY, Ren Y, Gong GY, Zong M, Liu F, Chen QJ, Xu Y (2018). Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep 37, 1353-1356.
DOI URL |
| [85] |
Veillet F, Perrot L, Chauvin L, Kermarrec MP, Guyon- Debast A, Chauvin JE, Nogué F, Mazier M (2019). Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/ Cas9 cytidine base editor. Int J Mol Sci 20, 402.
DOI URL |
| [86] |
Veillet F, Perrot L, Guyon-Debast A, Kermarrec MP, Chauvin L, Chauvin JE, Gallois JL, Mazier M, Nogué F (2020). Expanding the CRISPR toolbox in P. patens using SpCas9-NG variant and application for gene and base editing in Solanaceae crops. Int J Mol Sci 21, 1024.
DOI URL |
| [87] |
Walton RT, Christie KA, Whittaker MN, Kleinstiver BP (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290-296.
DOI URL |
| [88] |
Wang SX, Zong Y, Lin QP, Zhang HW, Chai ZZ, Zhang DD, Chen KL, Qiu JL, Gao CX (2020). Precise, predictable multi-nucleotide deletions in rice and wheat using APOBEC-Cas9. Nat Biotechnol 38, 1460-1465.
DOI URL |
| [89] |
Wei C, Wang C, Jia M, Guo HX, Luo PY, Wang MG, Zhu JK, Zhang H (2021). Efficient generation of homozygous substitutions in rice in one generation utilizing an rABE8e base editor. J Integr Plant Biol 63, 1595-1599.
DOI URL |
| [90] |
Wu J, Chen C, Xian GY, Liu DX, Lin L, Yin SL, Sun QF, Fang YJ, Zhang H, Wang YP (2020). Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol J 18, 1857-1859.
DOI URL |
| [91] |
Xie JK, Huang XY, Wang X, Gou SX, Liang YH, Chen FB, Li N, Ouyang Z, Zhang QJ, Ge WK, Jin Q, Shi H, Zhuang ZP, Zhao XZ, Lian M, Wang JW, Ye YH, Quan LQ, Wu H, Wang KP, Lai LX (2020). ACBE, a new base editor for simultaneous C-to-T and A-to-G substitutions in mammalian systems. BMC Biol 18, 131.
DOI URL |
| [92] |
Xu RF, Kong FN, Qin RY, Li J, Liu XS, Wei PC (2021). Development of an efficient plant dual cytosine and adenine editor. J Integr Plant Biol 63, 1600-1605.
DOI URL |
| [93] |
Xu RF, Li J, Liu XS, Shan TF, Qin RY, Wei PC (2020a). Development of plant prime-editing systems for precise genome editing. Plant Commun 1, 100043.
DOI URL |
| [94] |
Xu W, Zhang CW, Yang YX, Zhao S, Kang GT, He XQ, Song JL, Yang JX (2020b). Versatile nucleotides substitution in plant using an improved prime editing system. Mol Plant 13, 675-678.
DOI URL |
| [95] |
Yan F, Kuang YJ, Ren B, Wang JW, Zhang DW, Lin HH, Yang B, Zhou XP, Zhou HB (2018). Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant 11, 631-634.
DOI URL |
| [96] |
Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR (2018). In vivo base editing of post-mitotic sensory cells. Nat Commun 9, 2184.
DOI URL |
| [97] |
Zeng DC, Liu TL, Tan JT, Zhang YL, Zheng ZY, Wang B, Zhou DG, Xie XR, Guo MH, Liu YG, Zhu QL (2020). PhieCBEs: plant high-efficiency cytidine base editors with expanded target range. Mol Plant 13, 1666-1669.
DOI URL |
| [98] |
Zhang R, Liu JX, Chai ZZ, Chen S, Bai Y, Zong Y, Chen KL, Li JY, Jiang LJ, Gao CX (2019). Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat Plants 5, 480-485.
DOI PMID |
| [99] |
Zhang XH, Zhu BY, Chen L, Xie L, Yu WS, Wang Y, Li LX, Yin SM, Yang L, Hu HD, Han HH, Li YM, Wang LR, Chen G, Ma XY, Geng HQ, Huang WF, Pang XF, Yang ZZ, Wu YX, Siwko S, Kurita R, Nakamura Y, Yang L, Liu MY, Li DL (2020). Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat Biotechnol 38, 856-860.
DOI URL |
| [100] |
Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ (2013). Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell 50, 488-503.
DOI PMID |
| [101] |
Zhao DD, Li J, Li SW, Xin XQ, Hu MZ, Price MA, Rosser SJ, Bi CH, Zhang XL (2021). Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol 39, 35-40.
DOI URL |
| [102] |
Zhou CY, Sun YD, Yan R, Liu YJ, Zuo EW, Gu C, Han LX, Wei Y, Hu XD, Zeng R, Li YX, Zhou HB, Guo F, Yang H (2019). Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571, 275-278.
DOI URL |
| [103] | Zhu HC, Li C, Gao CX (2020). Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat Rev Mol Cell Biol 21, 661-677. |
| [104] |
Zhu XG, Zhu JK (2021). Precision genome editing heralds rapid de novo domestication for new crops. Cell 184, 1133-1134.
DOI URL |
| [105] |
Zong Y, Liu YJ, Xue CX, Li BS, Li XY, Wang YP, Li J, Liu GW, Huang XX, Cao XF, Gao CX (2022). An engineered prime editor with enhanced editing efficiency in plants. Nat Biotechnol doi: 10.1038/s41587-022-01254-w.
DOI |
| [106] | Zong Y, Song QN, Li C, Jin S, Zhang DB, Wang YP, Qiu JL, Gao CX (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol 36, 950-953. |
| [107] |
Zong Y, Wang YP, Li C, Zhang R, Chen KL, Ran YD, Qiu JL, Wang DW, Gao CX (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35, 438-440.
DOI URL |
| [108] |
Zou RS, Liu Y, Wu B, Ha T (2021). Cas9 deactivation with photocleavable guide RNAs. Mol Cell 81, 1553-1565.
DOI URL |
| [109] | Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018). De novo domestication of wild tomato using genome editing. Nat Biotechnol 81, 1211-1216. |
| [110] |
Zuo EW, Sun YD, Yuan TL, He BB, Zhou CY, Ying WQ, Liu J, Wei W, Zeng R, Li YX, Yang H (2020). A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat Methods 17, 600-604.
DOI URL |
| [1] | 周文期, 周玉乾, 李永生, 何海军, 杨彦忠, 王晓娟, 连晓荣, 刘忠祥, 胡筑兵. 玉米ZmICE2基因调控气孔发育[J]. 植物学报, 2023, 58(6): 866-881. |
| [2] | 苗春妍, 李铭铭, 左鑫, 丁宁, 杜家方, 李娟, 张重义, 王丰青. 湖北地黄CRISPR/Cas9基因编辑体系的建立[J]. 植物学报, 2023, 58(6): 905-916. |
| [3] | 邱锐, 何峰, 李瑞, 王亚梅, 邢思年, 曹英萍, 刘叶飞, 周昕越, 赵彦, 付春祥. 柳枝稷木质素基因F5H的高效编辑[J]. 植物学报, 2023, 58(2): 298-307. |
| [4] | 邓娴, 李彤, 曹晓风. 基因编辑在饲草育种中的应用与展望[J]. 植物学报, 2023, 58(2): 233-240. |
| [5] | 王娜, 姜腾, 王彬锡, 牛丽芳, 林浩. 单倍体育种技术研究进展及其在苜蓿等豆科牧草中的应用[J]. 植物学报, 2022, 57(6): 756-763. |
| [6] | 张瑞, 高彩霞. 基于双碱基编辑系统的植物基因靶向随机突变技术[J]. 植物学报, 2021, 56(1): 50-55. |
| [7] | 谢先荣, 曾栋昌, 谭健韬, 祝钦泷, 刘耀光. 基于CRISPR编辑系统的DNA片段删除技术[J]. 植物学报, 2021, 56(1): 44-49. |
| [8] | 苏钺凯,邱镜仁,张晗,宋振巧,王建华. CRISPR/Cas9系统在植物基因组编辑中技术改进与创新的研究进展[J]. 植物学报, 2019, 54(3): 385-395. |
| [9] | 王影, 李相敢, 邱丽娟. CRISPR/Cas9基因组定点编辑中脱靶现象的研究进展[J]. 植物学报, 2018, 53(4): 528-541. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||