

植物学报 ›› 2021, Vol. 56 ›› Issue (1): 33-43.DOI: 10.11983/CBB20147 cstr: 32102.14.CBB20147
张雅文1,2, 梁山1, 徐国云3, 郭无瑕2,4,*( ), 邓书林2,4,*(
), 邓书林2,4,*( )
)
收稿日期:2020-08-25
接受日期:2021-01-05
出版日期:2021-01-01
发布日期:2021-01-15
通讯作者:
郭无瑕,邓书林
作者简介:E-mail: guowx@scbg.ac.cn;基金资助:
Yawen Zhang1,2, Shan Liang1, Guoyun Xu3, Wuxia Guo2,4,*( ), Shulin Deng2,4,*(
), Shulin Deng2,4,*( )
)
Received:2020-08-25
Accepted:2021-01-05
Online:2021-01-01
Published:2021-01-15
Contact:
Wuxia Guo,Shulin Deng
摘要: 烟草(Nicotiana tabacum)是基因功能分析的模式植物以及重要的经济作物之一, 适宜的生存环境对烟草的生长和繁殖至关重要。COL (CONSTANS-like)基因家族编码蛋白不仅调控植物开花, 而且在植物生物/非生物胁迫响应中发挥重要作用。该研究通过鉴定烟草COL基因家族成员, 分析其基因结构、进化关系、转录调控元件和表达模式, 探究其编码蛋白的生物学功能, 尤其是在烟草响应低温胁迫中的可能作用。结果显示, 在烟草中共鉴定出15个COL基因, 其编码的蛋白理化性质相近; 进化分析结果表明其包括3类, 每个类别的成员之间具有相似的外显子/内含子结构以及motif数量和类型; 烟草COL基因启动子区域含有大量与光、低温、干旱以及植物激素等响应相关的顺式作用元件; 基于二代高通量测序分析结果表明, 低温显著影响烟草COL基因的表达, 但对不同基因的影响存在差异, 不同COL基因的亲本(林烟草(N. sylvestris) (母本)和绒毛状烟草(N. tomentosiformis) (父本))具有表达偏好性, 且这种偏好性大部分会从6-7叶期保持到现蕾期。
张雅文, 梁山, 徐国云, 郭无瑕, 邓书林. 烟草CONSTANS-like基因家族的鉴定与分析. 植物学报, 2021, 56(1): 33-43.
Yawen Zhang, Shan Liang, Guoyun Xu, Wuxia Guo, Shulin Deng. Genome-wide Identification and Analysis of CONSTANS-like Gene Family in Nicotiana tabacum. Chinese Bulletin of Botany, 2021, 56(1): 33-43.
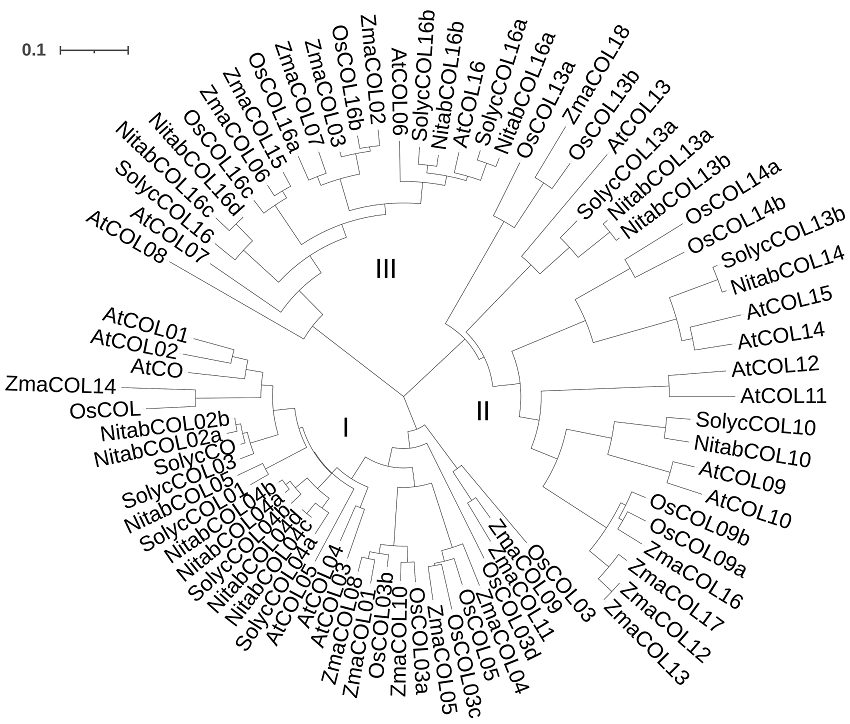
图1 5个物种COL蛋白的系统发育树 从NCBI数据库下载烟草(Nitab)、拟南芥(At)、番茄(Solyc)、水稻(Os)和玉米(Zma) COL蛋白序列。采用Clustal软件进行序列比对, 采用邻接法构建系统发育树, bar表示进化距离。
Figure 1 Phylogenetic tree of COL proteins from five species COL protein sequences of Nicotiana tabacum (Nitab), Arabidopsis thaliana (At), Solanum lycopersicum (Solyc), Oryza sativa (Os), and Zea mays (Zma) were downloaded from NCBI database. The phylogenetic tree was constructed by neighbor joining method with Clustal, bar indicated evolutionary distance.
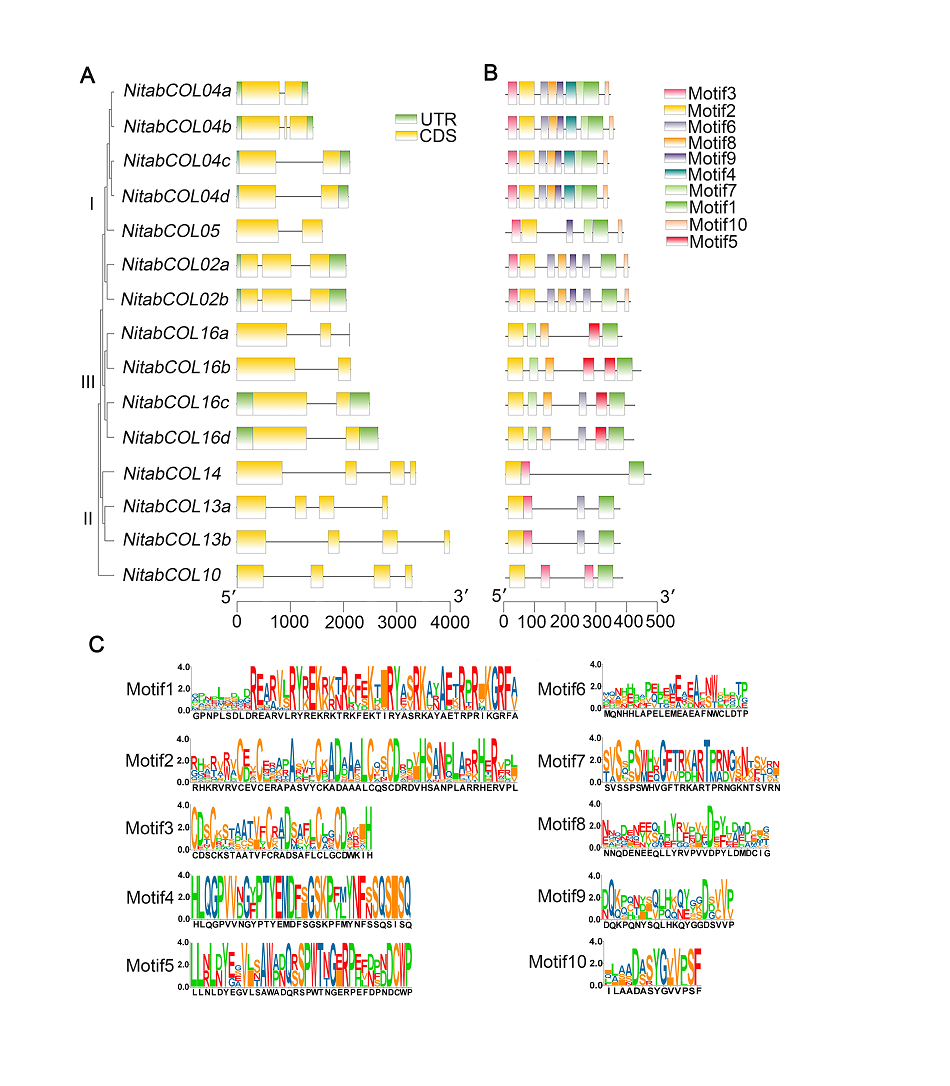
图2 烟草COL的基因结构(A)、COL蛋白的保守结构域(B)及蛋白保守基序的氨基酸序列(C) 不同颜色的矩形代表不同的蛋白保守基序以及非编码区(UTR) (绿色)和编码序列(CDS) (黄色)。比例尺分别代表基因序列长度(A)和蛋白序列长度(B), 纵坐标数字代表蛋白保守基序中相应氨基酸的频率(C)。
Figure 2 Gene structures of COL genes (A), and conserved motifs (B) and amino acid sequences (C) of conserved motifs of COL proteins in tobacco Differently colored rectangles representing different protein conserved motifs as well as untranslated region (UTR) (green) and coding sequence (CDS) (yellow). Scale bars indicating gene sequence length (A) and protein sequence length (B), respectively, and the ordinate numbers indicating the frequency of corresponding amino acid in the conserved motifs (C).
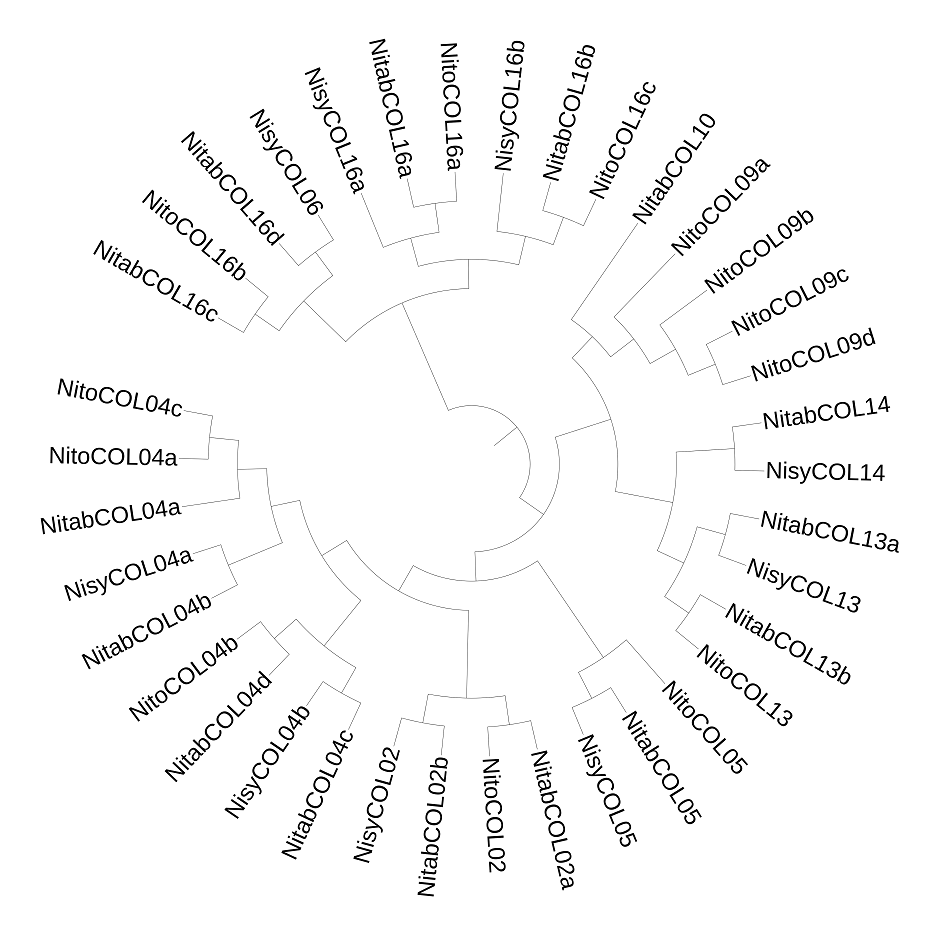
图3 烟草及其亲本COL蛋白的系统发育树 从茄科基因组数据库(Solgenomics.net)下载普通烟草(Nitab) (子代)、绒毛状烟草(Nito) (父本)、林烟草(Nisy) (母本) COL蛋白序列, 采用Clustal软件进行序列比对, 并用邻接法构建系统发育树。
Figure 3 Phylogenetic tree of the COL family proteins in tobacco and its parents The COL proteins of Nicotiana tabacum (Nitab) (progeny), N. tomentosiformis (Nito) (paternal), and N. sylvestris (Nisy) (maternal) were downloaded from Solgenomics.net. The phylogenetic tree was constructed by neighbor joining method with Clustal.
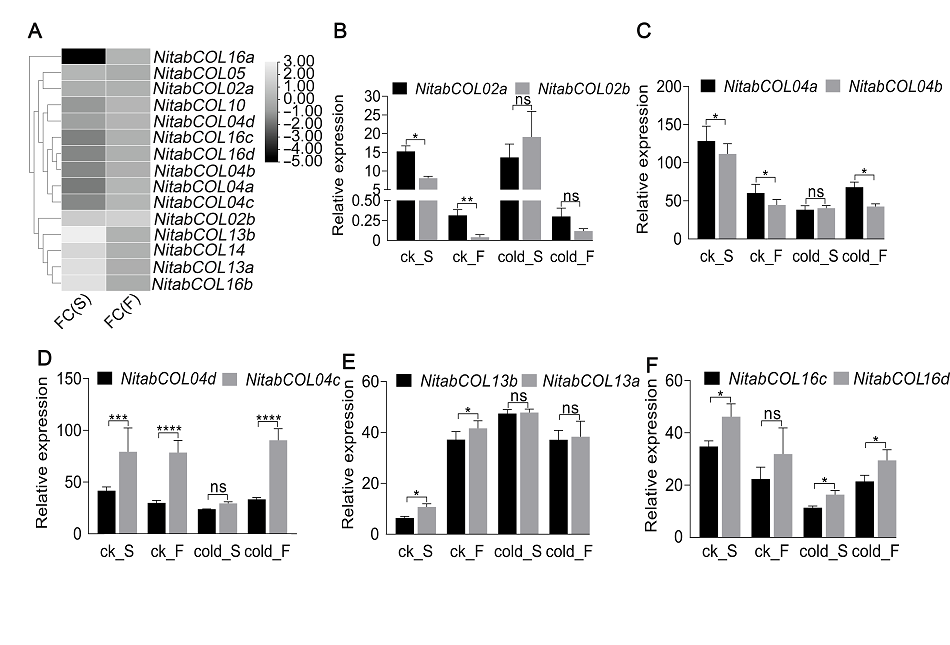
图4 烟草所有COL基因(A)和5对部分同源基因(B)-(F)在低温处理下不同发育阶段叶片中的表达 FC(S): 12°C处理下6-7叶期的表达差异倍数; FC(F): 12°C处理下现蕾期的表达差异倍数; ck_S: 26°C下6-7叶期烟草; ck_F: 26°C下现蕾期烟草; cold_S: 12°C处理后6-7叶期烟草; cold_F: 12°C处理后现蕾期烟草; 黑色: 来自父本的基因; 灰色: 来自母本的基因。 相对表达水平为3次生物学重复的平均值, 误差线代表标准差, 采用Student’s t-test对部分同源基因对进行差异显著性检验, * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001, ns表示无显著性差异。
Figure 4 Expression of all COL genes (A) and five homologous genes pairs (B)-(F) in tobacco leaves at different developmental stages under cold treatment FC(S): Fold change at 6-7 leaf stage under 12°C treatment; FC(F): Fold change at budding stage under 12°C treatment; ck_S: 6-7 leaf stage under 26°C; ck_F: Budding stage under 26°C; cold_S: 6-7 leaf stage under 12°C treatment; cold_F: Budding stage under 12°C treatment; Black: Orthologs from paternal plant; Gray: Orthologs from maternal plant; the relative expression level is the average of 3 biological replicates, the error bar represents standard deviation, and Student’s t-test is used for significance test, * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001, and ns means not significant.
| [1] | 樊希彬, 李丹, 左晓晴, 薛金燕 (2016). 气候条件对烟草生长的影响分析. 黑龙江农业科学 ( 4), 27-30. |
| [2] | 王玲, 郭长奎, 任丁, 马红 (2017). 水稻非生物胁迫响应基因OsMIP1的表达与进化分析. 植物学报 52, 43-53. |
| [3] |
Borden KLB (2000). RING domains: master builders of molecular scaffolds? J Mol Biol 295, 1103-1112.
DOI URL PMID |
| [4] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13, 1194-1202.
DOI URL PMID |
| [5] |
Crocco CD, Botto JF (2013). BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531, 44-52.
DOI URL PMID |
| [6] |
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010). Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61, 651-679.
URL PMID |
| [7] |
Datta S, Hettiarachchi GHCM, Deng XW, Holm M (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18, 70-84.
URL PMID |
| [8] |
Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF (2008). Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42, 443-461.
DOI URL PMID |
| [9] |
Flagel LE, Wendel JF (2010). Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol 186, 184-193.
DOI URL PMID |
| [10] |
Griffiths S, Dunford RP, Coupland G, Laurie DA (2003). The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131, 1855-1867.
DOI URL PMID |
| [11] | Holm M, Hardtke CS, Gaudet R, Deng XW (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20, 118-127. |
| [12] |
Hu TH, Wei QZ, Wang WH, Hu HJ, Mao WH, Zhu QM, Bao CL (2018). Genome-wide identification and characterization of CONSTANS-like gene family in radish (Raphanus sativus). PLoS One 13, e0204137.
URL PMID |
| [13] |
Jin JJ, Zhang H, Zhang JF, Liu PP, Chen X, Li ZF, Xu YL, Lu P, Cao PJ (2017). Integrated transcriptomics and metabolomics analysis to characterize cold stress responses in Nicotiana tabacum. BMC Genomics 18, 496.
DOI URL PMID |
| [14] |
Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21, 3416-3420.
DOI URL PMID |
| [15] |
Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR (2008). The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann Bot 101, 805-814.
DOI URL PMID |
| [16] | Li AL, Liu DC, Wu J, Zhao XB, Hao M, Geng SF, Yan J, Jiang XX, Zhang LQ, Wu JY, Yin LY, Zhang RZ, Wu L, Zheng YL, Mao L (2014). mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26, 1878-1900. |
| [17] |
Liao Y, Smyth GK, Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930.
URL PMID |
| [18] |
Liu H, Dong SY, Sun DY, Liu W, Gu FW, Liu YZ, Guo T, Wang H, Wang JF, Chen ZQ (2016a). CONSTANS-Like 9 (OsCOL9) interacts with receptor for activated C-Kinase 1 (OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS One 11, e0166249.
URL PMID |
| [19] |
Liu JH, Shen JQ, Xu Y, Li XH, Xiao JH, Xiong LZ (2016b). Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J Exp Bot 67, 5785-5798.
DOI URL PMID |
| [20] | Liu ZL, Adams KL (2007). Expression partitioning between genes duplicated by polyploidy under abiotic stress and during organ development. Curr Biol 17, 1669-1674. |
| [21] |
Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550.
URL PMID |
| [22] |
Min JH, Chung JS, Lee KH, Kim CS (2015). The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J Integr Plant Biol 57, 313-324.
DOI URL PMID |
| [23] |
Miura K, Furumoto T (2013). Cold signaling and cold response in plants. Int J Mol Sci 14, 5312-5337.
DOI URL PMID |
| [24] |
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847-857.
DOI URL PMID |
| [25] | Qin WQ, Yu Y, Jin YY, Wang XD, Liu J, Xi JP, Li Z, Li HQ, Zhao G, Hu W, Chen CJ, Li FQ, Yang ZE (2018). Genome-wide analysis elucidates the role of CONSTANS- like genes in stress responses of cotton. Int J Mol Sci 19, 2658. |
| [26] | Saidi Y, Finka A, Goloubinoff P (2011). Heat perception and signaling in plants: a tortuous path to thermotolerance. New Phytol 190, 556-565. |
| [27] | Sauter M (2013). Root responses to flooding. Curr Opin Plant Biol 16, 282-286. |
| [28] |
Sierro N, Battey JND, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014). The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5, 3833.
DOI URL PMID |
| [29] |
Singh D, Laxmi A (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 6, 895.
DOI URL PMID |
| [30] |
Skalická K, Lim KY, Matyasek R, Matzke M, Leitch AR, Kovarik A (2005). Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol 166, 291-303.
URL PMID |
| [31] |
Song YH, Song NY, Shin SY, Kim HJ, Yun DJ, Lim CO, Lee SY, Kang KY, Hong JC (2008). Isolation of CONSTANS as a TGA4/OBF4 interacting protein. Mol Cells 25, 559-565.
URL PMID |
| [32] |
Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116-1120.
DOI URL PMID |
| [33] |
Tang XL, Mu XM, Shao HB, Wang HY, Brestic M (2015). Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit Rev Biotechnol 35, 425-437.
DOI URL PMID |
| [34] |
Yamori W, Evans JR, Von Caemmerer S (2010). Effects of growth and measurement light intensities on temperature dependence of CO2 assimilation rate in tobacco leaves. Plant Cell Environ 33, 332-343.
DOI URL PMID |
| [35] |
Yoo MJ, Szadkowski E, Wendel JF (2013). Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110, 171-180.
DOI URL PMID |
| [36] |
Zhang TZ, Hu Y, Jiang WK, Fang L, Guan XY, Chen JD, Zhang JB, Saski CA, Scheffler BE, Stelly DM, Hulse- Kemp AM, Wan Q, Liu BL, Liu CX, Wang S, Pan MQ, Wang YK, Wang DW, Ye WX, Chang LJ, Zhang WP, Song QX, Kirkbride RC, Chen XY, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu XY, Zhang H, Wu HT, Zhou L, Mei GF, Chen SQ, Tian Y, Xiang D, Li XH, Ding J, Zuo QY, Tao LN, Liu YC, Li J, Lin Y, Hui YY, Cao ZS, Cai CP, Zhu XF, Jiang Z, Zhou BL, Guo WZ, Li RQ, Chen ZJ (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33, 531-537.
DOI URL PMID |
| [37] |
Zhang ZL, Ji RH, Li HY, Zhao T, Liu J, Lin CT, Liu B (2014). CONSTANS-LIKE 7 (COL7) is involved in phytochrome B (phyB)-mediated light-quality regulation of auxin homeostasis. Mol Plant 7, 1429-1440.
DOI URL PMID |
| [1] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [2] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [3] | 欧阳子龙, 贾湘璐, 石景忠, 滕维超, 刘秀. 生长调节剂对低温胁迫及复温下红海榄幼苗光合特性的影响[J]. 植物生态学报, 2025, 49(4): 638-652. |
| [4] | 樊蓓, 任敏, 王延峰, 党峰峰, 陈国梁, 程国亭, 杨金雨, 孙会茹. 番茄SlWRKY45转录因子在响应低温和干旱胁迫中的功能(长英文摘要)[J]. 植物学报, 2025, 60(2): 186-203. |
| [5] | 高敏, 缑倩倩, 王国华, 郭文婷, 张宇, 张妍. 低温胁迫对不同母树年龄柠条锦鸡儿种子萌发幼苗生理和生长的影响[J]. 植物生态学报, 2024, 48(2): 201-214. |
| [6] | 师生波, 周党卫, 李天才, 德科加, 杲秀珍, 马家麟, 孙涛, 王方琳. 青藏高原高山嵩草光合功能对模拟夜间低温的响应[J]. 植物生态学报, 2023, 47(3): 361-373. |
| [7] | 师生波, 师瑞, 周党卫, 张雯. 低温对高山嵩草叶片光化学和非光化学能量耗散特征的影响[J]. 植物生态学报, 2023, 47(10): 1441-1452. |
| [8] | 陈奕竹, 郎伟光, 陈效逑. 中国北方树木秋季物候的过程模拟及其区域分异归因[J]. 植物生态学报, 2022, 46(7): 753-765. |
| [9] | 于海英, 杨莉琳, 付素静, 张志敏, 姚琦馥. 暖温带森林木本植物展叶始期对低温和热量累积变化的响应[J]. 植物生态学报, 2022, 46(12): 1573-1584. |
| [10] | 吴丹丹, 陈永坤, 杨宇, 孔春艳, 龚明. 小桐子半胱氨酸蛋白酶家族和相应miRNAs的鉴定及其对低温锻炼的响应[J]. 植物学报, 2021, 56(5): 544-558. |
| [11] | 赵燕,周俭民. 萤火素酶互补实验检测蛋白互作[J]. 植物学报, 2020, 55(1): 69-75. |
| [12] | 杨小青,黄晓琴,韩晓阳,刘腾飞,岳晓伟,伊冉. 外源物质对茶树耐寒及蔗糖代谢关键基因表达的影响[J]. 植物学报, 2020, 55(1): 21-30. |
| [13] | 刘利静,赵庆臻,谢旗,于菲菲. 快速高效检测植物体内蛋白泛素化修饰研究方法[J]. 植物学报, 2019, 54(6): 753-763. |
| [14] | 刘栋峰,唐永严,雒胜韬,罗伟,李志涛,种康,徐云远. 利用低温水浴鉴定水稻苗期耐寒性[J]. 植物学报, 2019, 54(4): 509-514. |
| [15] | 陈建权, 程晨, 张梦恬, 张向前, 张尧, 王爱英, 祝建波. 天山雪莲SiSAD基因与拟南芥AtFAB2基因转化 烟草的抗寒性分析[J]. 植物学报, 2018, 53(5): 603-611. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||