

植物学报 ›› 2020, Vol. 55 ›› Issue (1): 76-82.DOI: 10.11983/CBB19208 cstr: 32102.14.CBB19208
收稿日期:2019-10-24
接受日期:2019-12-31
出版日期:2020-01-01
发布日期:2020-01-03
通讯作者:
任东涛
基金资助:
Dan Zhu,Hanwei Cao,Yuan Li,Dongtao Ren( )
)
Received:2019-10-24
Accepted:2019-12-31
Online:2020-01-01
Published:2020-01-03
Contact:
Dongtao Ren
摘要: 蛋白磷酸化是一种重要的蛋白质翻译后修饰方式, 几乎参与植物所有生命过程的调节。蛋白磷酸化过程主要指在蛋白激酶的催化作用下, 将三磷酸腺苷(ATP)上的γ位磷酸基团转移到底物蛋白特定氨基酸残基上的过程。底物蛋白上被磷酸化的常见氨基酸有丝氨酸、苏氨酸及酪氨酸, 磷酸基团与氨基酸中的羟基通过酯键连接。该文详细描述了几种常用的蛋白质体外及体内磷酸化的检测方法及注意事项。
朱丹,曹汉威,李媛,任东涛. 植物蛋白磷酸化的检测方法. 植物学报, 2020, 55(1): 76-82.
Dan Zhu,Hanwei Cao,Yuan Li,Dongtao Ren. Protocols for Analyzing Plant Phospho-proteins. Chinese Bulletin of Botany, 2020, 55(1): 76-82.
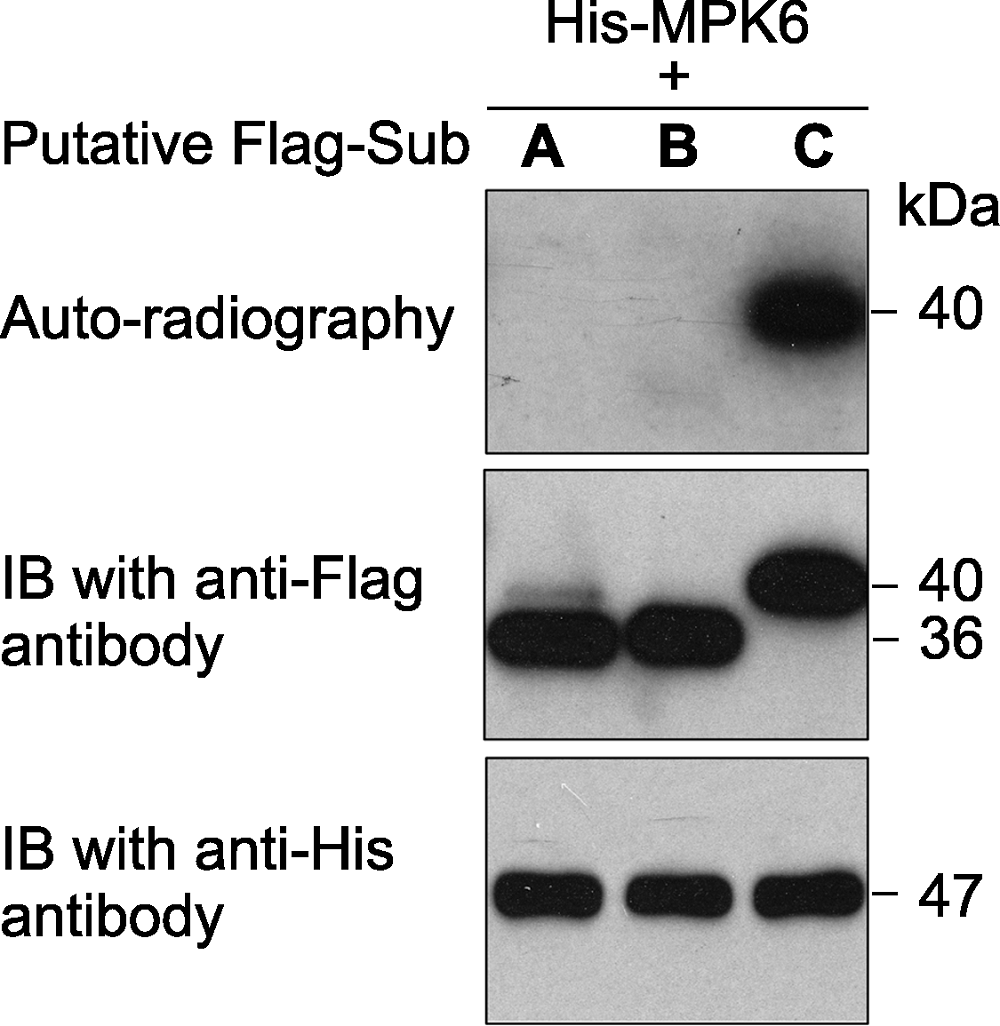
图1 原核表达的拟南芥激酶MPK6对3个推测底物的体外磷酸分析 激酶(MPK6)连接有6×His标签、推测的底物蛋白(A、B和C)连接有Flag标签。体外磷酸化反应完成后, 反应体系中的蛋白用SDS-PAGE gel分离。然后进行磷酸化底物的放射自显影分析(上图)。底物蛋白(中图)和激酶(下图)的免疫印迹分析显示底物、激酶各自在磷酸化反应中的蛋白使用量基本一致。
Figure 1 An in vitro phosphorylation assay of 3 putative substrate proteins by MPK6 in Arabidopsis thaliana MPK6 was fused with the 6×His tag and the 3 putative substrates were fused with the Flag tag. After phosphorylation reaction, the proteins in the mixture were separated on a SDS- PAGE gel. The gel was dried and exposed to X-ray film (top). Immunoblotting with anti-His and anti-Flag antibodies were used to show the levels of the substrate (middle) and kinase (bottom) proteins in the reactions.
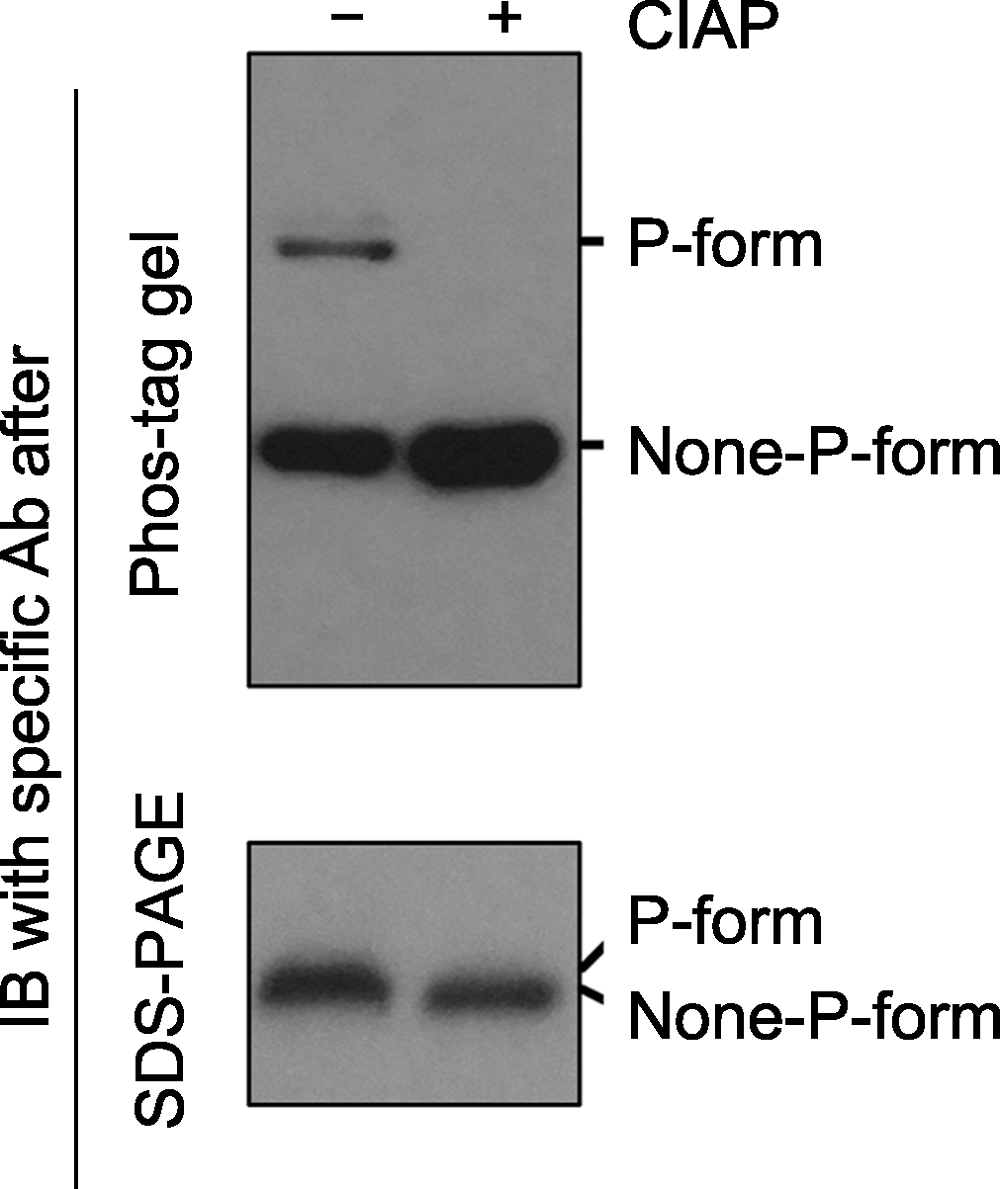
图2 Phos-tag gel和SDS-PAGE gel电泳后, 免疫印迹检测磷酸化蛋白的对比分析 磷酸酶(如Calf intestine alkaline phosphatase, CIAP)可去除磷酸化蛋白上的磷酸基团。同一样品在CIAP处理前(-)、后(+), 磷酸化和非磷酸化形式的蛋白经Phos-tag gel (上图)和SDS- PAGE gel (下图)电泳后的免疫印迹分析结果。经Phos-tag gel电泳, 未加CIAP的样品中特定蛋白的磷酸化(P-form)和非磷酸化形式(None-P-form)被清晰地分开(上图); 加CIAP后磷酸化形式消失而非磷酸化形式条带增强。经SDS-PAGE gel电泳, 未加和加CIAP处理的样品中, 特定蛋白的磷酸化和非磷酸化形式分开不明显(下图)。
Figure 2 Immunoblotting detection of phospho-proteins in samples after Phos-tag gel and SDS-PAGE gel separation Phospho-proteins can be dephosphorylated by phosphatases (e.g. Calf intestine alkaline phosphatase, CIAP). Protein samples treated with (+) or without (-) CIAP were separated by Phos-tag (top) and SDS-PAGE (bottom) gels, and the specific protein was further detected by immunoblotting. The P-form and None-P-form of the protein were separated clearly by a Phos-tag gel (top), while the two forms were not separated by a SDS-PAGE gel (bottom). The missing of the upper band and the increasing of the bottom band after Phos-tag gel separation indicated that P-form protein was completely dephosphorylated by CIAP treatment.
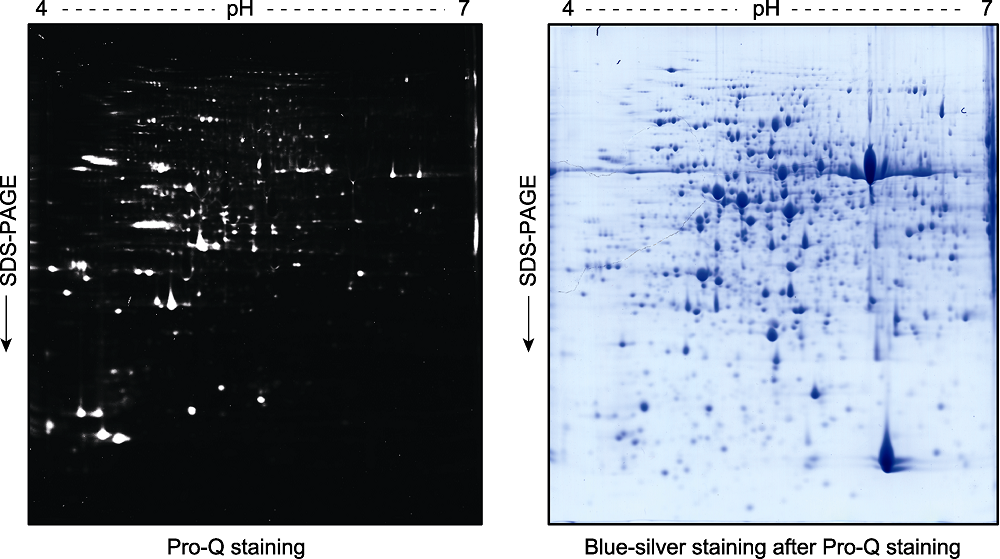
图3 拟南芥幼苗总蛋白双向凝胶电泳后的磷酸化蛋白Pro-Q Diamond染色检测分析 2周龄拟南芥幼苗总蛋白提取液经双向凝胶电泳分离后, 用Pro-Q Diamond试剂对磷酸化蛋白进行染色(左图)。荧光扫描后的凝胶再进行考马斯亮蓝染色(blue-silver staining) (右图)。
Figure 3 Pro-Q staining assay of the phospho-proteins in total proteins extracted from Arabidopsis thaliana seedlings and separated by a 2D gel Two-week-old Arabidopsis thaliana seedlings were used for total protein extraction. The total proteins were separated by a 2D gel and the phospho-proteins were stained by Pro-Q (left). The gel was scanned with a Typhoon 9410 fluorescence scanner, and then stained with blue-silver to visualize the proteins (right).
| [1] | Agrawal GK, Thelen JJ (2005). Development of a simplified, economical polyacrylamide gel staining protocol for phosphoproteins. Proteomics 5, 4684-4688. |
| [2] | Agrawal GK, Thelen JJ (2006). Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol Cell Proteomics 5, 2044-2059. |
| [3] | Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633-1649. |
| [4] | Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG (2004). Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327-1333. |
| [5] | Chao Q, Liu XY, Mei YC, Gao ZF, Chen YB, Qian CR, Hao YB, Wang BC (2014). Light-regulated phosphorylation of maize phosphoenolpyruvate carboxykinase plays a vital role in its activity. Plant Mol Biol 85, 95-105. |
| [6] | Chen MJ, Dixon JE, Manning G (2017). Genomics and evolution of protein phosphatases. Sci Signal 10, eaag1796. |
| [7] | de la Fuente van Bentem S, Hirt H (2007). Using phosphoproteomics to reveal signaling dynamics in plants. Trends Plant Sci 12, 404-411. |
| [8] | Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM (2002). Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol 20, 301-305. |
| [9] | Fischer EH, Krebs EG (1955). Conversion of phosphorylase B to phosphorylase A in muscle extracts. J Biol Chem 216, 121-132. |
| [10] | Frost DC, Li LJ (2014). Recent advances in mass spectrometry-based glycoproteomics. Adv Protein Chem Struct Biol 95, 71-123. |
| [11] | Hubbard MJ, Cohen P (1993). On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem Sci 18, 172-177. |
| [12] | Ke YQ, Han GQ, He HQ, Li JX (2009). Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem Biophys Res Commun 379, 133-138. |
| [13] | Khan M, Takasaki H, Komatsu S (2005). Comprehensive phosphoproteome analysis in rice and identification of phosphoproteins responsive to different hormones/stresses. J Proteome Res 4, 1592-1599. |
| [14] | Kim HS, Fernandes G, Lee CW (2016). Protein phosphatases involved in regulating mitosis: facts and hypotheses. Mol Cell 39, 654-662. |
| [15] | Kinoshita E, Kinoshita-Kikuta E, Koike T (2007). Specific recognition and detection of phosphorylated proteins using characteristics of metal ion. Yakugaku Zasshi 127, 1897-1913. |
| [16] | Kosako H, Nagano K (2011). Quantitative phosphoproteomics strategies for understanding protein kinase-mediated signal transduction pathways. Expert Rev Proteomics 8, 81-94. |
| [17] | Krupa A, Preethi G, Srinivasan N (2004). Structural modes of stabilization of permissive phosphorylation sites in protein kinases: distinct strategies in Ser/Thr and Tyr kinases. J Mol Biol 339, 1025-1039. |
| [18] | Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. |
| [19] | Laugesen S, Bergoin A, Rossignol M (2004). Deciphering the plant phosphoproteome: tools and strategies for a challenging task. Plant Physiol Biochem 42, 929-936. |
| [20] | Peck SC (2003). Early phosphorylation events in biotic stress. Curr Opin Plant Biol 6, 334-338. |
| [21] | Prak S, Hem S, Boudet J, Viennois G, Sommerer N, Rossignol M, Maurel C, Santoni V (2008). Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics 7, 1019-1030. |
| [22] | Wang PC, Zhao Y, Li ZP, Hsu CC, Liu X, Fu LW, Hou YJ, Du YY, Xie SJ, Zhang CG, Gao JH, Cao MJ, Huang XS, Zhu YF, Tang K, Wang XG, Tao WA, Xiong Y, Zhu JK (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell 69, 100-112. |
| [23] | Whiteman SA, Nühse TS, Ashford DA, Sanders D, Maathuis FJ (2008). A proteomic and phosphoproteomic analysis of Oryza sativa plasma membrane and vacuolar membrane. Plant J 56, 146-156. |
| [24] | Wu CF, Wang RN, Liang QJ, Liang JJ, Li WK, Jung SY, Qin J, Lin SH, Kuang J (2010). Dissecting the M phase- specific phosphorylation of serine-proline or threonine- proline motifs. Mol Biol Cell 21, 1470-1481. |
| [25] | Yang C, Wang ZG, Zhu PF (2004). Recent advances of protein phosphorylation in proteome. Prog Physiol Sci 35, 119-124. |
| [26] | Yin XJ, Wang X, Komatsu S (2018). Phosphoproteomics: protein phosphorylation in regulation of seed germination and plant growth. Curr Protein Pept Sci 19, 401-412. |
| [27] | Zhang X, Cui YN, Yu M, Su BD, Gong W, Baluška F, Komis G, Samaj J, Shan XY, Lin JX (2019). Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol 181, 480-498. |
| [28] | Zhu WG (2017). Regulation of p53 acetylation. Sci China Life Sci 60, 321-323. |
| [1] | 陈龙 郭柯 勾晓华 赵秀海 马泓若. 祁连圆柏林群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 0-0. |
| [2] | 惠城阳, 章巧依, 刘腾腾, 刘维勇, 周丽娜, 金鑫杰, 张永华, 刘金亮. 温州大罗山主要植被类型及物种组成特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [3] | 曹毅 张松林 王旭峰 杨安昌 任敏慧 杨浩 韩超. 兰州市南北两山植物群落数据集[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [4] | 童金莲, 张博纳, 汤璐瑶, 叶琳峰, 李姝雯, 谢江波, 李彦, 王忠媛. C4植物狗尾草功能性状网络沿降水梯度带的区域分异规律[J]. 植物生态学报, 2025, 49(预发表): 1-. |
| [5] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [6] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [7] | 赵珮杉 高广磊 丁国栋 张英. 林龄和生态位对樟子松人工林地下真菌群落构建的影响[J]. 植物生态学报, 2025, 49(地上地下生态过程关联): 1-0. |
| [8] | 张子睿, 周静, 胡艳萍, 梁爽, 马永鹏, 陈伟乐. 极度濒危植物巧家五针松的根内和根际真菌群落特征[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [9] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [10] | 黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群野生植物荔波杜鹃保护遗传学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [11] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [12] | 高雨轩, 苏艳军, 冯育才, 张军, 汪小全, 刘玲莉. 珍稀濒危孑遗植物银杉的研究与保护现状[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [13] | 贾妍妍, 柳华清, 解欣然, 王博, 张维, 杨允菲. 珍稀濒危植物天山梣林龄结构及种群动态[J]. 植物生态学报, 2025, 49(5): 760-772. |
| [14] | 平晓燕, 杜毅倩, 赖仕蓉, 孔梦桥, 余国杰. 植物应对食草动物采食的化学防御策略研究进展[J]. 植物生态学报, 2025, 49(5): 667-680. |
| [15] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||