

植物学报 ›› 2019, Vol. 54 ›› Issue (3): 343-349.DOI: 10.11983/CBB18106 cstr: 32102.14.CBB18106
收稿日期:2018-04-26
接受日期:2018-12-10
出版日期:2019-05-01
发布日期:2019-11-24
通讯作者:
包颖
基金资助:
Xia Zhang1,Xiang Jing1,Guangcai Zhou2,Ying Bao1,*( )
)
Received:2018-04-26
Accepted:2018-12-10
Online:2019-05-01
Published:2019-11-24
Contact:
Ying Bao
摘要: 淀粉作为主要的碳水化合物在储藏能量方面发挥至关重要的作用。颗粒结合型淀粉合酶(GBSS)与直链淀粉的合成息息相关。尽管该酶的编码基因已在许多栽培植物中被分离和确定, 但有关它们在作物野生近缘种中的序列分歧和表达的研究却相对较少。该研究以药用野生稻(Oryza officinalis)为研究对象, 定性和定量地分析了GBSS编码基因的序列特点、与其它植物同源基因的进化关系以及在叶和种子中的表达情况。系统发育分析表明, 该酶在禾本科植物中分别由GBSSI和GBSSII基因编码。在药用野生稻中, 这2种基因所编码蛋白的氨基酸序列一致性为62%, 并且它们在不同器官内呈现时空分化表达, 其中GBSSI在种子中超强表达, GBSSII则主要在叶片表达。
张霞,景翔,周光才,包颖. 药用野生稻GBSS基因的系统发育及组织特异性表达. 植物学报, 2019, 54(3): 343-349.
Xia Zhang,Xiang Jing,Guangcai Zhou,Ying Bao. Phylogeny and Tissue-specific Expression of the GBSS Genes in Oryza officinalis. Chinese Bulletin of Botany, 2019, 54(3): 343-349.
| Gene | Primer name | Primer sequence (5'-3') |
|---|---|---|
| GBSSI | GBSSI-F | AACGTGGCTGCTCCTTGAA |
| GBSSI-R | TTGGCAATAAGCCACACACA | |
| GBSSII | GBSSII-F | AGGCATCGAGGGTGAGGAG |
| GBSSII-R | CCATCTGGCCCACATCTCTA |
表1 GBSSI和GBSSII的引物序列
Table 1 Primer sequences of GBSSI and GBSSII
| Gene | Primer name | Primer sequence (5'-3') |
|---|---|---|
| GBSSI | GBSSI-F | AACGTGGCTGCTCCTTGAA |
| GBSSI-R | TTGGCAATAAGCCACACACA | |
| GBSSII | GBSSII-F | AGGCATCGAGGGTGAGGAG |
| GBSSII-R | CCATCTGGCCCACATCTCTA |
| Plants | Gene (GenBank/UniProt No.) | Sequence identity (%) | |
|---|---|---|---|
| Oryza officinalis | |||
| GBSSI | GBSSII | ||
| Oryza sativa subsp. japonica | GBSSI (XP_025882300.1) | 95.22 | 60.35 |
| GBSSII (XP_015647210.1) | 62.30 | 96.72 | |
| O. sativa subsp. indica | GBSSI (AAN77100.1) | 97.87 | 62.30 |
| GBSSII (ACY56079.1) | 62.46 | 97.04 | |
| Leersia perrieri | GBSSI (A0A0D9WLF6) * | 93.01 | 62.15 |
| GBSSII (A0A0D9WED4) * | 58.44 | 85.62 | |
| Zizania latifolia | GBSSI (ASSH01051725.1) ** | 90.80 | 62.01 |
| GBSSII (ASSH01023543.1) ** | 59.46 | 87.47 | |
| Brachypodium distachyon | GBSSI (XP_003557139.1) | 79.31 | 61.16 |
| GBSSII (XP_003569238.1) | 60.33 | 85.43 | |
| Hordeum vulgare | GBSSI (BAC41202.1) | 81.97 | 60.76 |
| GBSSII (BAJ99426.1) | 61.85 | 86.12 | |
| Setaria italica | GBSSI (AGW27658.1) | 86.72 | 61.90 |
| GBSSII (XP_004956034.1) | 62.40 | 87.34 | |
| Sorghum bicolor | GBSSI (XP_002436418.1) | 82.52 | 61.90 |
| GBSSII (XP_002461889.1) | 62.62 | 86.18 | |
| Triticum aestivum | GBSSI (XP_020146905.1) | 81.37 | 60.36 |
| GBSSII (AAG27624.1) | 61.79 | 86.09 | |
| Zea mays | GBSSI (NP_001105001) | 82.08 | 62.13 |
| GBSSII (NP_001334833) | 62.36 | 85.88 | |
| Musa acuminata | GBSS1 (KF512020.1) | 66.39 | 65.35 |
| GBSS2 (KF512021.1) | 67.96 | 63.99 | |
| GBSS3 (KF512023.1) | 63.93 | 62.91 | |
| Arabidopsis thaliana | GBSS (XP_020866584.1) | 62.48 | 63.88 |
| Gossypium raimondii | GBSS1 (XP_012474755.1) | 63.89 | 66.01 |
| GBSS2 (XP_012439861.1) | 63.11 | 66.77 | |
| GBSS3 (XP_012486622.1) | 62.75 | 64.01 | |
| Solanum lycopersicum | GBSS (NP_001311457.1) | 67.05 | 62.03 |
| Carica papaya | GBSS (XP_021900468.1) | 65.91 | 66.45 |
| S. tuberosum | GBSS (XP_006343763.1) | 62.89 | 67.27 |
| Vitis vinifera | GBSS1 (XP_010660257.1) | 65.52 | 66.23 |
| GBSS2 (XP_019081062.1) | 64.07 | 67.76 | |
| Amborella trichopoda | GBSS (XP_006837847.1) | 62.81 | 66.17 |
| Chlamydomonas reinhardtii | GBSS (XP_001697117.1) | 47.79 | 47.05 |
表2 药用野生稻和其它植物的GBSS氨基酸序列一致度
Table 2 Amino acid sequence identities of GBSSs between Oryza officinalis and other plants
| Plants | Gene (GenBank/UniProt No.) | Sequence identity (%) | |
|---|---|---|---|
| Oryza officinalis | |||
| GBSSI | GBSSII | ||
| Oryza sativa subsp. japonica | GBSSI (XP_025882300.1) | 95.22 | 60.35 |
| GBSSII (XP_015647210.1) | 62.30 | 96.72 | |
| O. sativa subsp. indica | GBSSI (AAN77100.1) | 97.87 | 62.30 |
| GBSSII (ACY56079.1) | 62.46 | 97.04 | |
| Leersia perrieri | GBSSI (A0A0D9WLF6) * | 93.01 | 62.15 |
| GBSSII (A0A0D9WED4) * | 58.44 | 85.62 | |
| Zizania latifolia | GBSSI (ASSH01051725.1) ** | 90.80 | 62.01 |
| GBSSII (ASSH01023543.1) ** | 59.46 | 87.47 | |
| Brachypodium distachyon | GBSSI (XP_003557139.1) | 79.31 | 61.16 |
| GBSSII (XP_003569238.1) | 60.33 | 85.43 | |
| Hordeum vulgare | GBSSI (BAC41202.1) | 81.97 | 60.76 |
| GBSSII (BAJ99426.1) | 61.85 | 86.12 | |
| Setaria italica | GBSSI (AGW27658.1) | 86.72 | 61.90 |
| GBSSII (XP_004956034.1) | 62.40 | 87.34 | |
| Sorghum bicolor | GBSSI (XP_002436418.1) | 82.52 | 61.90 |
| GBSSII (XP_002461889.1) | 62.62 | 86.18 | |
| Triticum aestivum | GBSSI (XP_020146905.1) | 81.37 | 60.36 |
| GBSSII (AAG27624.1) | 61.79 | 86.09 | |
| Zea mays | GBSSI (NP_001105001) | 82.08 | 62.13 |
| GBSSII (NP_001334833) | 62.36 | 85.88 | |
| Musa acuminata | GBSS1 (KF512020.1) | 66.39 | 65.35 |
| GBSS2 (KF512021.1) | 67.96 | 63.99 | |
| GBSS3 (KF512023.1) | 63.93 | 62.91 | |
| Arabidopsis thaliana | GBSS (XP_020866584.1) | 62.48 | 63.88 |
| Gossypium raimondii | GBSS1 (XP_012474755.1) | 63.89 | 66.01 |
| GBSS2 (XP_012439861.1) | 63.11 | 66.77 | |
| GBSS3 (XP_012486622.1) | 62.75 | 64.01 | |
| Solanum lycopersicum | GBSS (NP_001311457.1) | 67.05 | 62.03 |
| Carica papaya | GBSS (XP_021900468.1) | 65.91 | 66.45 |
| S. tuberosum | GBSS (XP_006343763.1) | 62.89 | 67.27 |
| Vitis vinifera | GBSS1 (XP_010660257.1) | 65.52 | 66.23 |
| GBSS2 (XP_019081062.1) | 64.07 | 67.76 | |
| Amborella trichopoda | GBSS (XP_006837847.1) | 62.81 | 66.17 |
| Chlamydomonas reinhardtii | GBSS (XP_001697117.1) | 47.79 | 47.05 |
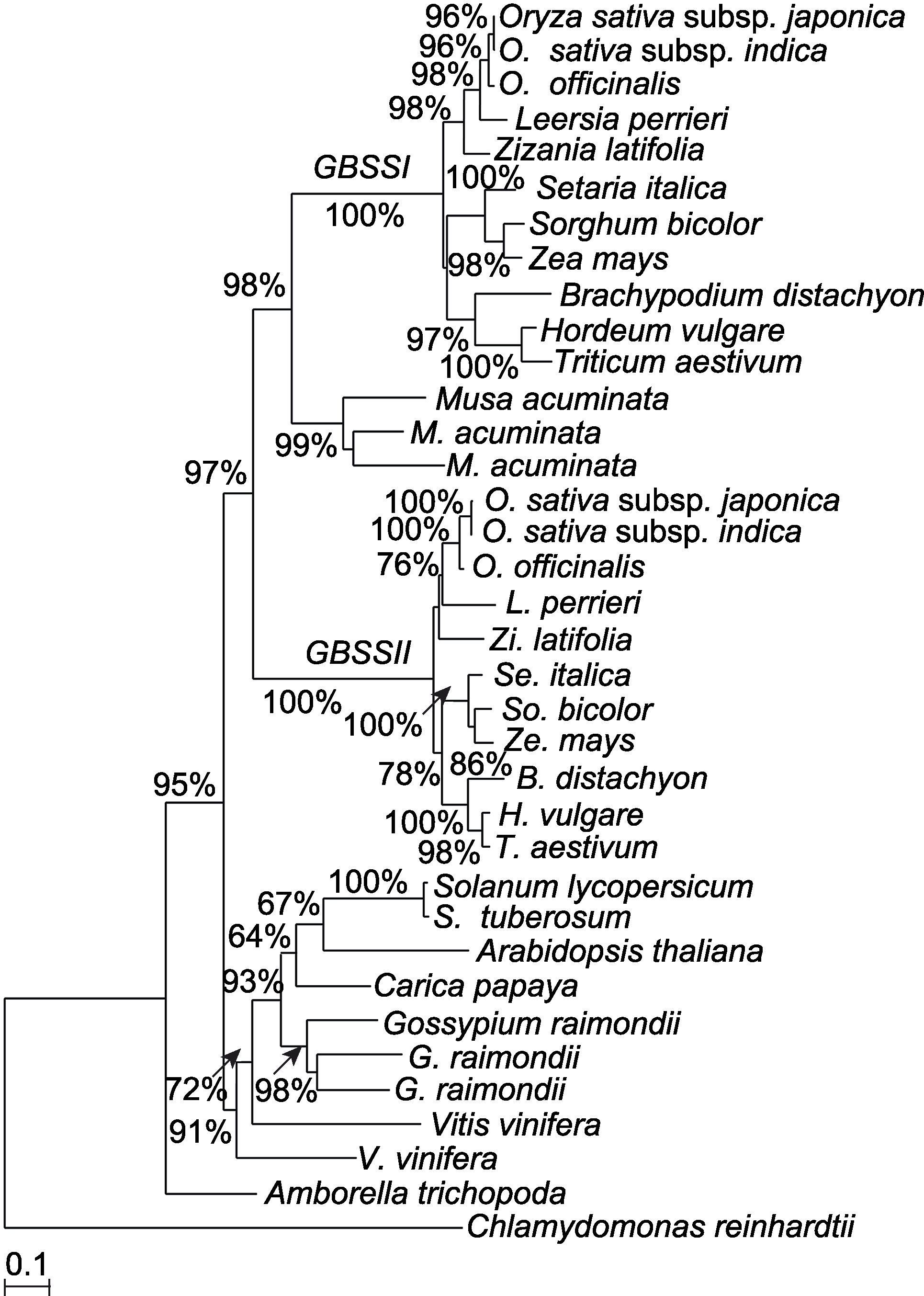
图2 利用氨基酸序列构建的淀粉合酶的最大似然性系统发育树分支旁的数字代表自展支持率。
Figure 2 A maximum likelihood phylogenetic tree of the GBSS based on amino acid sequences Numbers near branches indicate bootstrap value.
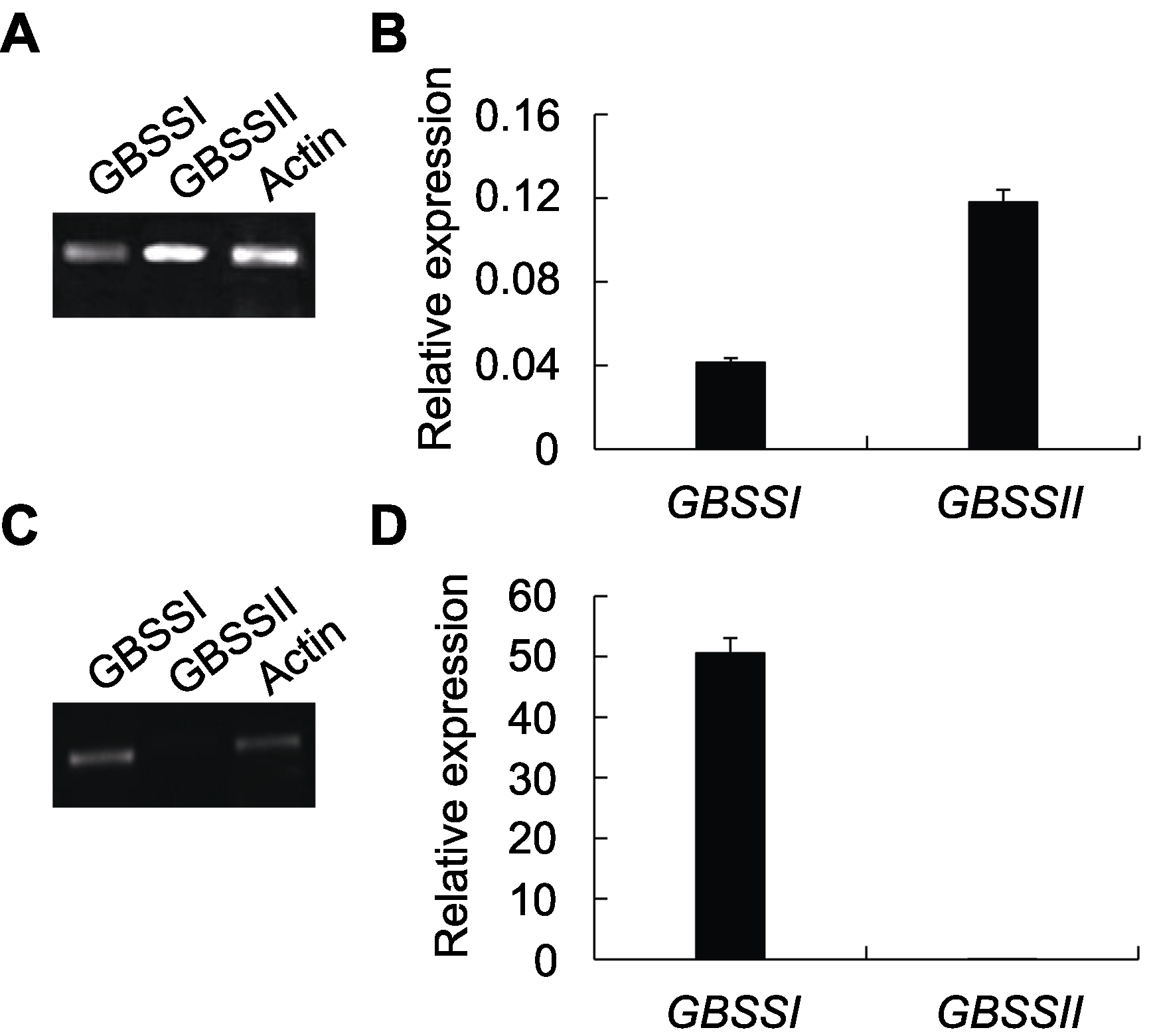
图3 GBSSI和GBSSII在药用野生稻叶和种子中的相对表达(A), (B) GBSS基因在叶中的RT-PCR和qRT-PCR扩增结果; (C), (D) GBSS基因在种子中的RT-PCR和qRT-PCR扩增结果。
Figure 3 Relative expression of GBSSI and GBSSII in leaves and seeds of Oryza officinalis(A), (B) Amplification results of RT-PCR and qRT-PCR for GBSS genes in leaves; (C), (D) Amplification results of RT-PCR and qRT-PCR for GBSS genes in seeds.
| [1] |
包颖, 杜家潇, 景翔, 徐思 (2015). 药用野生稻叶中淀粉合成酶基因家族的序列分化和特异表达. 植物学报 50, 683-690.
DOI URL |
| [2] |
陈凤花, 王琳, 胡丽华 (2005). 实时荧光定量RT-PCR内参基因的选择. 临床检验杂志 23, 393-395.
DOI URL |
| [3] |
顾燕娟 (2006). 支链淀粉合成相关基因等位基因间的差异对稻米淀粉理化特性的影响. 硕士论文. 扬州: 扬州大学. pp. 3-8.
DOI URL |
| [4] |
王倩, 孙文静, 包颖 (2017). 植物颗粒结合淀粉合酶GBSS基因家族的进化. 植物学报 52, 179-187.
DOI URL |
| [5] |
杨学明 (2003). 几个重复序列在不同稻种中的分布及其与稻种分化关系的研究. 硕士论文. 扬州: 扬州大学. pp. 6-10.
DOI URL |
| [6] |
张鹏 (2008). 抑制淀粉分支酶类基因表达对稻米品质影响的研究. 硕士论文. 扬州: 扬州大学. pp. 3-8.
DOI URL |
| [7] | Bao Y, Xu S, Jing X, Meng L, Qin ZY (2015). De novo assembly and characterization ofOryza officinalis leaf transcriptome by using RNA-seq. Biomed Res Int 2015, 982065. |
| [8] |
Cheng J, Khan MA, Qiu WM, Li J, Zhou H, Zhang Q, Guo WW, Zhu TT, Peng JH, Sun FJ, Li SH, Korban SS, Han YP (2012). Diversification of genes encoding granulebound starch synthase in monocots and dicots is marked by multiple genome-wide duplication events.PLoS One 7, e30088.
DOI URL PMID |
| [9] |
Dian WM, Jiang HW, Wu P (2005). Evolution and expression analysis of starch synthase III and IV in rice.J Exp Bot 56, 623-632.
DOI URL PMID |
| [10] |
Gouy M, Guindon S, Gascuel O (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building.Mol Biol Evol 27, 221-224.
DOI URL PMID |
| [11] |
Guindon S, Gascuel O (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood.Syst Biol 52, 696-704.
DOI URL PMID |
| [12] |
Ohdan T, Francisco Jr PB, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot 56, 3229-3244.
DOI URL PMID |
| [13] | Shure M, Wessler S, Fedoroff N (1983). Molecular identification and isolation of thewaxy locus in maize. Cell 35, 225-233. |
| [14] | Van Harsselaar JK, Lorenz J, Senning M, Sonnewald U, Sonnewald S (2017). Genome-wide analysis of starch metabolism genes in potato (Solanum tuberosum L.). BMC Genomics 18, 37. |
| [15] |
Vrinten PL, Nakamura T (2000). Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues.Plant Physiol 122, 255-264.
DOI URL |
| [16] | Wang ZY, Wu ZL, Xing YY, Zheng FG, Guo XL, Zhang WG, Hong MM (1990). Nucleotide sequence of ricewaxy gene. Nucleic Acids Res 18, 5898. |
| [1] | 何斌 韩鹏宾 肖书礼 裴康迪 唐勤. 铁坚油杉林的群落类型及特征[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [2] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [3] | 刘世忠 张倩媚 张德强 刘菊秀 褚国伟 李跃林. 1999-2015年鼎湖山季风常绿阔叶林长期监测样地的植物物种组成和群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 1-0. |
| [4] | 白帆, 王杨. 2007–2015年北京东灵山暖温带落叶阔叶林长期监测样地植物物种组成和群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 1-. |
| [5] | 蔡榕榕, 沈历都, 刘雅各, 费雯丽, 戴冠华. 2005-2010年长白山阔叶红松林长期监测样地的物种组成和群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 1-. |
| [6] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [7] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [8] | 唐远翔, 熊仕臣, 朱洪锋, 张新生, 游成铭, 刘思凝, 谭波, 徐振锋. 长期氮添加对四川盆地西缘常绿阔叶林优势树种凋落叶产量及碳氮磷归还的影响[J]. 植物生态学报, 2025, 49(5): 720-731. |
| [9] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| [10] | 王贝贝, 吴苏, 王苗苗, 胡锦涛. 日光诱导叶绿素荧光不同组分在作物总初级生产力估算中的贡献比例: 多时间尺度分析[J]. 植物生态学报, 2025, 49(4): 562-572. |
| [11] | 李琳, 黄佳芳, 丁中浩, 郭萍萍, 蔡芫镔, 李诗华, 李云琴, 罗敏. 淹水高度增加对短叶茳芏潮汐湿地净生态系统CO2交换量的影响[J]. 植物生态学报, 2025, 49(4): 526-539. |
| [12] | 闫小莉, 刘贵梅, 李小玉, 江宇翔, 全小强, 王燕茹, 曲鲁平, 汤行昊. 不同氮添加水平和铵硝态氮配比环境下木荷幼苗光合及叶绿素荧光特性[J]. 植物生态学报, 2025, 49(4): 624-637. |
| [13] | 赵白龙, 李业亮, 王宇飞, 孙斌. 十大功劳属(小檗科)的叶结构分型新体系[J]. 植物学报, 2025, 60(4): 1-0. |
| [14] | 王传永, 庄典, 宋正达, 翟恒华, 李乃伟, 张凡. 黑果腺肋花楸叶绿体全基因组的结构和比较分析及系统进化推断[J]. 植物学报, 2025, 60(4): 1-0. |
| [15] | 杜英杰, 范爱连, 王雪, 闫晓俊, 陈廷廷, 贾林巧, 姜琦, 陈光水. 亚热带天然常绿阔叶林乔木树种与林下灌木树种根-叶功能性状协调性及差异[J]. 植物生态学报, 2025, 49(4): 585-595. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||