

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (4): 497-502.DOI: 10.11983/CBB19108 cstr: 32102.14.CBB19108
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Lichao Chen,Ni Zhan,Yansha Li,Jian Feng,Jianru Zuo*( )
)
Received:2019-06-13
Accepted:2019-06-20
Online:2019-07-01
Published:2020-01-08
Contact:
Jianru Zuo
Lichao Chen,Ni Zhan,Yansha Li,Jian Feng,Jianru Zuo. Detection and Analysis of Protein S-nitrosylation in Plants[J]. Chinese Bulletin of Botany, 2019, 54(4): 497-502.
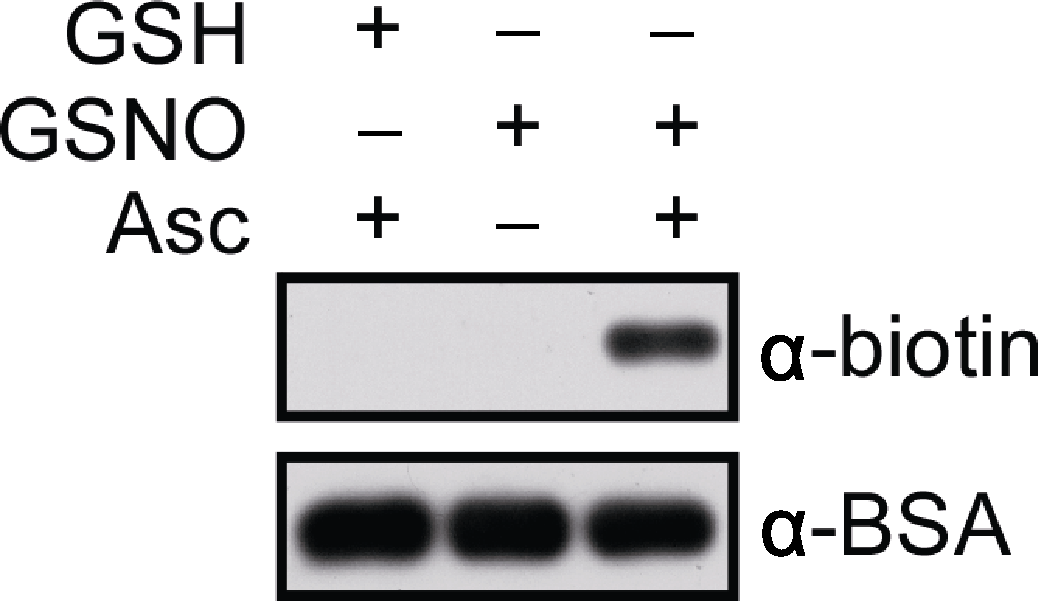
Figure 1 In vitro analysis of S-nitrosylated bovine serum albumin (BSA) BSA is labelled with biotin-maleimide, subjected to SDS- PAGE and western blotting. Anti-biotin and anti-BSA antibodies are used as primary antibodies, respectively (dilution in 1:20 000) and anti-mouse IgG as a secondary antibody. Signals are detected by using the SuperSignal Western Femto Maximun Sensitivity Substrate Kit. The blots are exposed for 20 sec (top) and 30 sec (bottom), respectively. GSH: Glutathione; GSNO: S-nitrosoglutathione; Asc: Sodium ascorbate
| [1] | Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sánchez-Vicente I, Nambara E, Lorenzo O ( 2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat Commun 6, 8669. |
| [2] | Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J, Zuo J ( 2009). The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res 19, 1377. |
| [3] | Cui B, Pan Q, Clarke D, Villarreal MO, Umbreen S, Yuan B, Shan W, Jiang J, Loake GJ ( 2018). S-nitrosylation of the zinc finger protein SRG1 regulates plant immunity. Nat Commun 9, 4226. |
| [4] | Feng J, Chen L, Zuo J ( 2019). Protein S-nitrosylation in plants: current progresses and challenges. J Integr Plant Biol doi. org/10.1111/jipb.12780. |
| [5] | Feng J, Wang C, Chen Q, Chen H, Ren B, Li X, Zuo J ( 2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat Commun 4, 1529. |
| [6] | He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, Fiorani F, Jackson RB, Crawford NM, Pei ZM ( 2004). Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968-1971. |
| [7] | Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS ( 2005). Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6, 150-166. |
| [8] | Hess DT, Stamler JS ( 2012). Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem 287, 4411-4418. |
| [9] | Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J ( 2015). Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol 167, 1731-1746. |
| [10] | Hu J, Yang H, Mu J, Lu T, Peng J, Deng X, Kong Z, Bao S, Cao X, Zuo J ( 2017). Nitric oxide regulates protein methylation during stress responses in plants. Mol Cell 67, 702-710. |
| [11] | Iglesias MJ, Terrile MC, Correa-Aragunde N, Colman SL, Izquierdo-Álvarez A, Fiol DF, París R, Sánchez-López N, Marina A, Calderón Villalobos LIA, Estelle M, Lamattina L, Martínez-Ruiz A, Casalongué CA ( 2018). Regulation of SCFTIR1/AFBs E3 ligase assembly by S-nitrosylation of Arabidopsis SKP1-like1 impacts on auxin signaling. Redox Biol 18, 200-210. |
| [12] | Jaffrey SR, Snyder SH ( 2001). The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001,pl1. |
| [13] | Ling T, Bellin D, Vandelle E, Imanifard Z, Delledonne M ( 2017). Host-mediated S-nitrosylation disarms the bacterial effector HopAI1 to reestablish immunity. Plant Cell 29, 2871-2881. |
| [14] | Lytvyn DI, Raynaud C, Yemets AI, Bergounioux C, Blume YB ( 2016). Involvement of inositol biosynthesis and nitric oxide in the mediation of UV-B induced oxidative stress. Front Plant Sci 7, 430. |
| [15] | Pan QN, Geng CC, Li DD, Xu SW, Mao DD, Umbreen S, Loake GJ, Cui BM ( 2019). Nitrate reductase-mediated nitric oxide regulates the leaf shape in Arabidopsis by mediating the homeostasis of reactive oxygen species. Int J Mol Sci 20, 2235. |
| [16] | París R, Vazquez MM, Graziano M, Terrile MC, Miller ND, Spalding EP, Otegui MS, Casalongué CA ( 2018). Distribution of endogenous NO regulates early gravitropic response and PIN2 localization in Arabidopsis roots. Front Plant Sci 9, 495. |
| [17] | Seth D, Hess DT, Hausladen A, Wang L, Wang YJ, Stamler JS ( 2018). A multiplex enzymatic machinery for cellular protein S-nitrosylation. Mol Cell 69, 451-464. |
| [18] | Seth P, Hsieh PN, Jamal S, Wang L, Gygi SP, Jain MK, Coller J, Stamler JS ( 2019). Regulation of microRNA machinery and development by interspecies S-nitrosylation. Cell 176, 1014-1025. |
| [19] | Shekariesfahlan A, Lindermayr C ( 2018). Identification of NO-sensitive cysteine residues using cysteine mutants of recombinant proteins. In: Mengel A, Lindermayr C, eds. Nitric Oxide: Methods and Protocols. New York: Springer New York. pp. 183-203. |
| [20] | Shi H, Liu W, Wei Y, Ye T ( 2017). Integration of auxin/ indole-3-acetic acid 17 and RGA-LIKE3 confers salt stress resistance through stabilization by nitric oxide in Arabidopsis. J Exp Bot 68, 1239-1249. |
| [21] | Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X ( 2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952-956. |
| [22] | Wang P, Zhu JK, Lang Z ( 2015). Nitric oxide suppresses the inhibitory effect of abscisic acid on seed germination by S-nitrosylation of SnRK2 proteins. Plant Signal Behav 10, e1031939. |
| [23] | Willems P, Horne A, Van Parys T, Goormachtig S, De Smet I, Botzki A, Van Breusegem F, Gevaert K ( 2019). The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J 99, 752-762. |
| [24] | Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J ( 2010). GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS One 5, e11290. |
| [25] | Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou JM, Zuo J ( 2015). S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol 167, 1604. |
| [26] | Zhan N, Wang C, Chen L, Yang H, Feng J, Gong X, Ren B, Wu R, Mu J, Li Y, Liu Z, Zhou Y, Peng J, Wang K, Huang X, Xiao S, Zuo J ( 2018). S-nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol Cell 71, 142-154. |
| [27] | Zhang L, Shi X, Zhang Y, Wang J, Yang J, Ishida T, Jiang W, Han X, Kang J, Wang X, Pan L, Lv S, Cao B, Zhang Y, Wu J, Han H, Hu Z, Cui L, Sawa S, He J, Wang G ( 2019 a). CLE9 peptide-induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana. Plant Cell Environ 42, 1033-1044. |
| [28] | Zhang ZW, Fu YF, Zhou YH, Wang CQ, Lan T, Chen GD, Zeng J, Chen YE, Yuan M, Yuan S, Hu JY ( 2019 b). Nitrogen and nitric oxide regulate Arabidopsis flowering differently. Plant Sci 284, 177-184. |
| [1] | Lu Zi-Jia, Wang Tian-Rui, Zheng Si-Si, Cao Jian-Guo, Kozlowski Gregor, Song Yi-Gang. Environmental adaptive genetic variation and genetic vulnerability of relict plant Pterocarya hupehensis [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | ZHANG Zi-Rui, Zhou Jing, HU Yan-Ping, Liang Shuang, MA Yong-Peng, CHEN Wei-Le. Root-associated Fungal Communities of the Critically Endangered Plant Pinus Squamata [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [3] | Tao Xie, Yifan Zhang, Yunhui Liu, Huiyu You, Jibenben Xia, Rong Ma, Chunni Zhang, Xuejun Hua. Research progress of iron-sulfur cluster synthesis system and regulation in plant mitochondria [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [4] | JIANG Xiao-Yu, YU Xin-Miao, LIAO Qin, ZHANG Jin-Wei, WU Xue-Feng, WANG Xu, PAN Jun-Tong, WANG Jun-Feng, MU Chun-Sheng, SHI Yu-Jie. Studies on the emission of nitrous oxide from terrestrial plants [J]. Chin J Plant Ecol, 2025, 49(4): 513-525. |
| [5] | Gan Xie, Jing Xuan, Qidi Fu, Ze Wei, Kai Xue, Hairui Luo, Jixi Gao, Min Li. Establishing an intelligent identification model for unmanned aerial vehicle surveys of grassland plant diversity [J]. Biodiv Sci, 2025, 33(4): 24236-. |
| [6] | Xiong Lianglin, Liang Guolu, Guo Qigao, Jing Danlong. Advances in the Regulation of Alternative Splicing of Genes in Plants in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2025, 60(3): 435-448. |
| [7] | Zhang Chan, Zhao Suya, Zhang Xinran, Wang Yifan, Wang Linlin. Impacts of alien pollinators on native plant‒pollinator interactions [J]. Biodiv Sci, 2025, 33(2): 24443-. |
| [8] | Yan Deng, Limin Lu, Qiang Zhang, Zhiduan Chen, Haihua Hu. A Comprehensive Evaluation of the Plastid DNA Data Gaps of Vascular Plants in Species and Geographic Area [J]. Chinese Bulletin of Botany, 2025, 60(1): 1-16. |
| [9] | Jing Xuan, Qidi Fu, Gan Xie, Kai Xue, Hairui Luo, Ze Wei, Mingyue Zhao, Liang Zhi, Huawei Wan, Jixi Gao, Min Li. An Artificial Intelligence Model for Identifying Grassland Plants in Northern China [J]. Chinese Bulletin of Botany, 2025, 60(1): 74-80. |
| [10] | Ruoyue Li, Xiaochao Yang, Zhanqing Hao, Shihong Jia. The intensity of heat waves and insect herbivory on campus plants and their relationship with leaf functional traits [J]. Biodiv Sci, 2025, 33(1): 24283-. |
| [11] | Xiangtan Yao, Xinyi Zhang, Yang Chen, Ye Yuan, Wangda Cheng, Tianrui Wang, Yingxiong Qiu. Genomic resequencing reveals the genetic diversity of the cultivated water caltrop, and the origin and domestication of ‘Nanhuling’ [J]. Biodiv Sci, 2024, 32(9): 24212-. |
| [12] | WANG Yi-Tong, Yeerjiang BAIKETUERHAN, LIAO Dan, WANG Juan. Correlation between elemental biometric characteristics and sexual dimorphism in leaves of dioecious Acer barbinerve at different growth stages [J]. Chin J Plant Ecol, 2024, 48(6): 760-769. |
| [13] | Chunjiao Xia, Yunguang Li, Shu Xia, Wei Pang, Chunli Chen. Flow Cytometric Analysis and Sorting in Plant Genomics [J]. Chinese Bulletin of Botany, 2024, 59(5): 774-782. |
| [14] | QU Ze-Kun, ZHU Li-Qin, JIANG Qi, WANG Xiao-Hong, YAO Xiao-Dong, CAI Shi-Feng, LUO Su-Zhen, sCHEN Guang-Shui. Nutrient foraging strategies of arbuscular mycorrhizal tree species in a subtropical evergreen broadleaf forest and their relationship with fine root morphology [J]. Chin J Plant Ecol, 2024, 48(4): 416-427. |
| [15] | Lansha Luo, Wenpei Song, Qingzhu Hua, Dawei Li, Hong Liang, Xianzhi Zhang. Research Progress on Plant Sex-determination Genes and Their Epigenetic Regulation [J]. Chinese Bulletin of Botany, 2024, 59(2): 278-290. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||