

Chinese Bulletin of Botany ›› 2017, Vol. 52 ›› Issue (5): 579-589.DOI: 10.11983/CBB16158 cstr: 32102.14.CBB16158
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Yan Yang1,2, Haiqin Zhang1, Xing Fan1, Lina Sha1, Houyang Kang1, Yi Wang1, Yonghong Zhou1*
Received:2016-07-31
Accepted:2016-11-11
Online:2017-09-01
Published:2017-07-10
Contact:
Yonghong Zhou
Yan Yang, Haiqin Zhang, Xing Fan, Lina Sha, Houyang Kang, Yi Wang, Yonghong Zhou. Polymorphism of Gliadin and Glutelin and Systematics Studies in Elytrigia[J]. Chinese Bulletin of Botany, 2017, 52(5): 579-589.
| No. | Species | Genome | 2n | Accession No. | Origin |
|---|---|---|---|---|---|
| 1 | Elytrigia bessarabica (Savul & Rayss) Dubovik | E | 2x | PI531711 | Estonia |
| 2 | Et. caespitosa ssp. nodosa (Nevski) Tzvel | ESt | 4x | PI531734 | Estonia |
| 3 | Et. caespitosa (C. Koch) Nevski | ESt | 4x | PI547311 | Leningrad Russian Federation |
| 4 | Et. elongata (Host) Nevski | E | 2x | PI578682 | Nebraska United States |
| 5 | Et. farcta (Viv.) Holub | EEE | 6x | PI531727 | France |
| 6 | Et. geniculata (Nevski) Tzvelev | StSt | 4x | PI639753 | Russian Federation |
| 7 | Et. geniculata ssp. pruinifera (Nevski) Tzvelev | - | 6x | PI547374 | Russian Federation |
| 8 | Et. intermedia (Host) Nevski | EESt | 6x | PI223668 | Iran |
| 9 | Et. intermedia ssp. intermedia (Host) Nevski | EESt | 6x | PI229917 | Iran |
| 10 | Et. kosaninii (Nábélek) Dubovik | St | 2x | PI237636 | Turkey |
| 11 | Et. libanotica (Hackel) Holub | St | 2x | PI229583 | Iran |
| 12 | Et. pachynera Prokudin | - | 6x | PI440059 | Former Soviet Union |
| 13 | Et. podpera (Nábělek) Holub | EEE | 6x | PI228387 | Iran |
| 14 | Et. pontica (Podp.) Holub | EEEEE | 10x | PI636523 | Argentina |
| 15 | Et. pungens (Pers.) Tutin | EStStP | 8x | PI547268 | Leningrad Russian Federation |
| 16 | Et. pycnantha (Godr.) Á. Löve | EStP | 6x | PI531741 | France |
| 17 | Et. rechingeri (Runemark) Hulub | EE | 4x | PI531745 | Greece |
| 18 | Et. repens (L.) Nevski | StStH | 6x | Y0814 | China |
| 19 | Et. scirpea (K. Presl) Holub | EE | 4x | PI531749 | Italy |
| 20 | Et. scythica (Nevski) Nevski | ESt | 4x | PI283272 | Former Soviet Union |
| 21 | Et. stipifolia (Czern. ex Nevski) Nevski | St | 2x | PI313960 | Former Soviet Union |
| 22 | Et. strigosa (M. Bieb.) Nevski | StSt | 4x | PI531752 | Estonia |
| 23 | Et. tauri (Boiss. & Bal.) Tzvel | St | 2x | PI401330 | Iran |
| 24 | Et. varnensis (Velen.) Holub | EEEEStSt | 12x | PI281863 | Germany |
Table 1 The materials of Elytrigia used in this study
| No. | Species | Genome | 2n | Accession No. | Origin |
|---|---|---|---|---|---|
| 1 | Elytrigia bessarabica (Savul & Rayss) Dubovik | E | 2x | PI531711 | Estonia |
| 2 | Et. caespitosa ssp. nodosa (Nevski) Tzvel | ESt | 4x | PI531734 | Estonia |
| 3 | Et. caespitosa (C. Koch) Nevski | ESt | 4x | PI547311 | Leningrad Russian Federation |
| 4 | Et. elongata (Host) Nevski | E | 2x | PI578682 | Nebraska United States |
| 5 | Et. farcta (Viv.) Holub | EEE | 6x | PI531727 | France |
| 6 | Et. geniculata (Nevski) Tzvelev | StSt | 4x | PI639753 | Russian Federation |
| 7 | Et. geniculata ssp. pruinifera (Nevski) Tzvelev | - | 6x | PI547374 | Russian Federation |
| 8 | Et. intermedia (Host) Nevski | EESt | 6x | PI223668 | Iran |
| 9 | Et. intermedia ssp. intermedia (Host) Nevski | EESt | 6x | PI229917 | Iran |
| 10 | Et. kosaninii (Nábélek) Dubovik | St | 2x | PI237636 | Turkey |
| 11 | Et. libanotica (Hackel) Holub | St | 2x | PI229583 | Iran |
| 12 | Et. pachynera Prokudin | - | 6x | PI440059 | Former Soviet Union |
| 13 | Et. podpera (Nábělek) Holub | EEE | 6x | PI228387 | Iran |
| 14 | Et. pontica (Podp.) Holub | EEEEE | 10x | PI636523 | Argentina |
| 15 | Et. pungens (Pers.) Tutin | EStStP | 8x | PI547268 | Leningrad Russian Federation |
| 16 | Et. pycnantha (Godr.) Á. Löve | EStP | 6x | PI531741 | France |
| 17 | Et. rechingeri (Runemark) Hulub | EE | 4x | PI531745 | Greece |
| 18 | Et. repens (L.) Nevski | StStH | 6x | Y0814 | China |
| 19 | Et. scirpea (K. Presl) Holub | EE | 4x | PI531749 | Italy |
| 20 | Et. scythica (Nevski) Nevski | ESt | 4x | PI283272 | Former Soviet Union |
| 21 | Et. stipifolia (Czern. ex Nevski) Nevski | St | 2x | PI313960 | Former Soviet Union |
| 22 | Et. strigosa (M. Bieb.) Nevski | StSt | 4x | PI531752 | Estonia |
| 23 | Et. tauri (Boiss. & Bal.) Tzvel | St | 2x | PI401330 | Iran |
| 24 | Et. varnensis (Velen.) Holub | EEEEStSt | 12x | PI281863 | Germany |
| No. | The num- ber of gliadin bands | The num- ber of glutelin bands | No. | The number of gliadin bands | The number of glutelin bands |
|---|---|---|---|---|---|
| 1 | 14 | 14 | 13 | 22 | 20 |
| 2 | 19 | 14 | 14 | 31 | 22 |
| 3 | 14 | 14 | 15 | 25 | 15 |
| 4 | 23 | 16 | 16 | 30 | 10 |
| 5 | 15 | 18 | 17 | 16 | 16 |
| 6 | 20 | 20 | 18 | 19 | 17 |
| 7 | 22 | 17 | 19 | 22 | 16 |
| 8 | 24 | 19 | 20 | 28 | 17 |
| 9 | 22 | 14 | 21 | 12 | 16 |
| 10 | 20 | 15 | 22 | 19 | 19 |
| 11 | 16 | 13 | 23 | 17 | 15 |
| 12 | 16 | 11 | 24 | 22 | 15 |
Table 2 The number of gliadin bands and glutelin bands in 24 accessions of Elytrigia
| No. | The num- ber of gliadin bands | The num- ber of glutelin bands | No. | The number of gliadin bands | The number of glutelin bands |
|---|---|---|---|---|---|
| 1 | 14 | 14 | 13 | 22 | 20 |
| 2 | 19 | 14 | 14 | 31 | 22 |
| 3 | 14 | 14 | 15 | 25 | 15 |
| 4 | 23 | 16 | 16 | 30 | 10 |
| 5 | 15 | 18 | 17 | 16 | 16 |
| 6 | 20 | 20 | 18 | 19 | 17 |
| 7 | 22 | 17 | 19 | 22 | 16 |
| 8 | 24 | 19 | 20 | 28 | 17 |
| 9 | 22 | 14 | 21 | 12 | 16 |
| 10 | 20 | 15 | 22 | 19 | 19 |
| 11 | 16 | 13 | 23 | 17 | 15 |
| 12 | 16 | 11 | 24 | 22 | 15 |
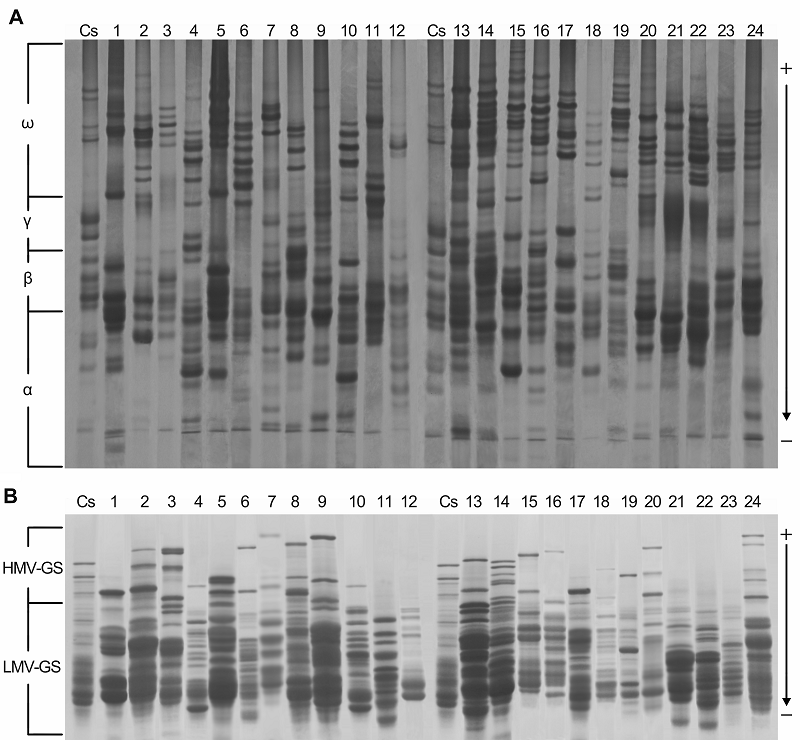
Figure 1 A-PAGE gliadin electyophoregram result (A) and SDS-PAGE glutelin electyophoregram result (B) of 24 accessions of ElytrigiaCs: China Spring. The accession number see Table 1.
| [1] | 陈默君, 贾慎修 (2002). 中国饲用植物. 北京: 中国农业出版社. pp. 136-140. |
| [2] | 董超华, 徐如宏, 张庆勤 (2003). 小麦醇溶蛋白和谷蛋白研究进展. 山地农业生物学报 22, 164-168. |
| [3] | 高梅, 张国权, 魏益民, 张继澍, 党娇, 王秋娟 (2007). 小麦醇溶蛋白A-PAGE电泳鉴定技术的分析. 西北农林科技大学学报(自然科学版) 35(9), 53-57. |
| [4] | 兰秀锦, 魏育明, 王志容, 郑有良 (1999). 中国节节麦与中东节节麦的醇溶蛋白遗传多样性比较研究. 四川农业大学学报 17, 245-248. |
| [5] | 李守明, 梁维, 魏凌基, 齐军仓, 王金玲 (2011). 利用两种电泳技术分析大麦品种的醇溶蛋白差异及亲缘关系. 石河子大学学报(自然科学版) 29, 15-19. |
| [6] | 李玉京, 李滨, 刘建中, 李继云, 李振声, 姚树江 (1998). 低磷营养胁迫对小麦-长穗偃麦草附加系酸性磷酸酶同工酶的影响. 中国农业科学 31(4), 26-31. |
| [7] | 吕伟东, 徐鹏彬, 蒲训 (2007). 偃麦草属种质资源在普通小麦育种中的应用现状简介. 草业学报 16, 136-140. |
| [8] | 马渐新, 周荣华, 董玉琛, 贾继增 (1999). 来自长穗偃麦草的抗小麦条锈病基因的定位. 科学通报 44, 65-69. |
| [9] | 孙善澄 (1981). 小偃麦新品种与中间类型的选育途径、程序和方法. 作物学报 7, 51-58. |
| [10] | 唐慧慧, 丁毅, 胡耀军 (2002). 中国近缘野生大麦醇溶蛋白的遗传多态性研究. 武汉植物学研究 20, 251-257. |
| [11] | 唐朝晖, 刘守斌, 刘少翔, 孙玉, 张兰萍, 逯成芳, 孙善澄, 刘广田 (2004). 二倍体长穗偃麦草高分子量谷蛋白亚基多态性研究. 华北农学报 19, 34-36. |
| [12] | 王红, 王瑞, 陈新宏, 赵继新, 钟刚 (2008). 国内外普通小麦醇溶蛋白的遗传差异. 西北农业学报 17(6), 67-72. |
| [13] | 王洪刚, 刘树兵, 亓增军, 孔凡晶, 高居荣 (2000). 中间偃麦草在小麦遗传改良中的应用研究. 山东农业大学学报(自然科学版) 31, 333-336. |
| [14] | 王际睿, 颜泽洪, 魏育明, 郑有良 (2004). 长穗偃麦草y型高分子量谷蛋白基因的鉴定与分子克隆. 农业生物技术学报 12, 143-146. |
| [15] | 吴卫东, 陈梦竹, 陈凡国, 夏光敏 (2010). 长穗偃麦草与老芒麦中α-醇溶蛋白基因的分离与鉴定. 分子植物育种 8, 20-28. |
| [16] | 杨瑞武, 魏秀华, 周永红, 郑有良 (2004). 赖草属植物醇溶蛋白的遗传多态性. 云南植物研究 26, 103-110. |
| [17] | 杨瑞武, 周永红, 郑有良 (2000). 披碱草属的醇溶蛋白研究. 四川农业大学学报 18, 11-14. |
| [18] | 杨瑞武, 周永红, 郑有良, 胡超 (2001). 小麦族四个属模式种的醇溶蛋白分析. 广西植物 21, 239-242. |
| [19] | 于海清 (2007). 拟鹅观草属四倍体物种的分子细胞遗传学研究. 博士论文. 雅安: 四川农业大学. pp. 12-42. |
| [20] | 翟旭光, 潘志芬, 商闯, 邓光兵, 余懋群 (2009). 燕麦麦谷蛋白SDS-PAGE电泳分析. 西南农业学报 22, 24-28. |
| [21] | 张学勇, 杨欣明, 董玉琛 (1995). 醇溶蛋白电泳在小麦种质资源遗传分析中的应用. 中国农业科学 28(4), 25-32. |
| [22] | 张颖, 张利, 张海琴, 丁春邦, 周永红 (2006). 鹅观草属、披碱草属、猬草属和仲彬草属物种的醇溶蛋白分析. 四川农业大学学报 24, 256-262. |
| [23] | 郑青焕, 李晓萍, 郭超, 昝凯, 陈真真, 白宇浩, 刘洋, 赵继新, 武军, 王中华, 陈新宏 (2016). 21份印度小麦高分子谷蛋白亚基、醇溶蛋白及品质分析. 麦类作物学报 36, 62-68. |
| [24] | 朱振东, 贾继增 (2003). 小麦SSR标记的发展及应用. 遗传 25, 355-360. |
| [25] | Asfaw Z (2008). Variation in hordein polypeptide pattern within Ethiopian barley, Hordeum vulgare L. (Poaceae).Hereditas 110, 185-191. |
| [26] | Assadi M, Runemark H (1995). Hybridisation, genomic constitution and generic delimitation in Elymus s.l. (Poa- ceae: Triticeae).Plant Syst Evol 194, 189-205. |
| [27] | Desvaux (1810). Eustachys. Paris: Nouv Bull Sci Soc Phi- lom. pp. 190. |
| [28] | Dewey DR (1980). Morphological, cytological and taxonomic relationships between Agropyron repens and Agropyron elongatiforme (Gramineae).Syst Bot 51, 61-70. |
| [29] | Dewey DR (1984). The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In: Gustafson JP, ed. Gene Manipulation in Plant Improvement. New York: Plenum Press. pp. 209-279. |
| [30] | Draper SR (1987). ISTA variety committee report of the working group for biochemical tests for cultivar identification 1983-1986.Seed Sci Technol 15, 431-434. |
| [31] | Dvořák J, Kasarda DD, Dietler MD, Lew EJL, Anderson OD, Litts JC, Shewry PR (1986). Chromosomal location of seed storage protein genes in the genome of Elytrigia elongata.Can J Genet Cytol 28, 818-830. |
| [32] | Jaccard P (1908). Nouvelles researches sur la distribution florale.Bull Soc Vaud Sci Nat 44, 223-270. |
| [33] | Jiang GM, Dong M (2000). A comparative study on photosynthesis and water use efficiency between clonal and non-clonal plant species along the Northeast China Transect (NECT).Acta Bot Sin 42, 855-863. |
| [34] | Kawaura K, Mochida K, Ogihara Y (2005). Expression profile of two storage-protein gene families in hexaploid wheat revealed by large-scale analysis of expressed sequence tags.Plant Physiol 139, 1870-1880. |
| [35] | Kim TW, Kim JC, Fedak G, Son JH, Park KC, Kim NS (2010). Sequence variation in ITS spacers and 5.8S rDNA and relationship of E, St, P, Ns, Xm, and H genomes in the genera of Agropyron, Elytrigia, Leymus, Pascopyrum, Psathyrostachys, and Hordeum. Genes Genomics 32, 477-485. |
| [36] | Lawrence GJ, Shepherd KW (1981). Chromosomal location of genes controlling seed proteins in species related to wheat.Theor Appl Genet 59, 25-31. |
| [37] | Li HJ, Wang XM (2009). Thinopyrum ponticum and Th. intermedium: the promising source of resistance to fungal and viral diseases of wheat.J Genet Genomics 36, 557-565. |
| [38] | Li ZS, Li B, Tong YP (2008). The contribution of distant hybridization with decaploid Agropyron elongatum to wheat improvement in China.J Genet Genomics 35, 451-456. |
| [39] | Löve Á (1984). Conspectus of the Triticeae.Feddes Rep 95, 425-521. |
| [40] | Mao PS, Huang Y, Wang XG, Meng L, Mao PC, Zhang GF (2010). Cytological evaluation and karyotype analysis in plant germplasms of Elytrigia Desv.Agric Sci China 9, 1553-1560. |
| [41] | Mason-Gamer RJ, Orme NL, Anderson CM (2002). Phylogenetic analysis of North American Elymus and the mono- genomic Triticeae (Poaceae) using three chloroplast DNA data sets.Genome 45, 991-1002. |
| [42] | Metakovsky EV, Akhmedov MG, Sozinov AA (1986). Genetic analysis of gliadin-encoding genes reveals gene clusters as well as single remote genes.Theor Appl Ge- net 73, 278-285. |
| [43] | Nakamura H (2000). Allelic variation at high-molecular- weight glutenin subunit loci, Glu-A1, Glu-B1 and Glu-D1 in Japanese and Chinese hexaploid wheats.Euphytica 112, 187-193. |
| [44] | Seberg O, Frederiksen S (2008). A phylogenetic analysis of the monogenomic Triticeae (Poaceae) based on morpho- logy.Bot J Linn Soc 136, 75-97. |
| [45] | Stebbins Jr GL, Pun FT (1953). Artificial and natural hybrids in the Gramineae, tribe Hordeae. V. Diploid hybrids of Agropyron.Am J Bot 40, 444-449. |
| [46] | Tzvelev NN (1976). Triticeae Dum Poaceae URSS. Leningrad: Nauka Publishers Leningrad Section. pp. 181-203. |
| [1] | CHENG Ke-Xin, DU Yao, LI Kai-Hang, WANG Hao-Chen, YANG Yan, JIN Yi, HE Xiao-Qing. Genetic mechanism of interaction between maize and phyllospheric microbiome [J]. Chin J Plant Ecol, 2024, 48(2): 215-228. |
| [2] | Wei Heping, Lu Tao, Jia Qiwei, Deng Fei, Zhu Hao, Qi Zehua, Wang Yuxi, Ye Hanfei, Yin Wenjing, Fang Yuan, Mu Dan, Rao Yuchun. QTL Mapping of Candidate Genes for Heading Date in Rice [J]. Chinese Bulletin of Botany, 2022, 57(5): 588-595. |
| [3] | Yanan Zhang, Lei Huang, Jiabin Li, Lei Zhang, Zhenhua Dang. Identification and Development of Polymorphic Genic-SSRs in Tamarix ramosissima in Alxa Region Based on Transcriptome [J]. Chinese Bulletin of Botany, 2021, 56(4): 433-442. |
| [4] | Xi Zhang, Tianhang Qiu, Anan Wang, Huajian Zhou, Min Yuan, Li Li, Sulan Bai, Suxia Cui. Morphology and Genetic Diversity of Phragmites australis in Beijing [J]. Chinese Bulletin of Botany, 2020, 55(6): 693-704. |
| [5] | Gang Ren, En Li, Shiye Zhao, Yanqiong Jiang, Shasha Wang, Sixian Tang, Huijian Hu. Correlation between color polymorphism and the MC1R gene of Lanius schach [J]. Biodiv Sci, 2020, 28(6): 688-694. |
| [6] | Jianfeng Huang, Lang Li, Jie Li. Polymorphism of the Internal Transcribed Spacer of nrDNA in Cinnamomum Schaeffer (Lauraceae) [J]. Chinese Bulletin of Botany, 2016, 51(5): 609-619. |
| [7] | Wen Fan, Ying Xu, Ting Xu, Jing Xu, Takahiro Yonezawa, Jiyin Gao, Wenju Zhang. Intragenomic Polymorphism of the Internal Transcribed Spacer Region of Ribosomal DNA in Camellia hongkongensis (Theaceae) and Species Identification [J]. Chinese Bulletin of Botany, 2015, 50(2): 217-226. |
| [8] | WANG Na-Na, CHEN Ying, YING Jiao-Yan, GAO Yong-Sheng, BAI Yong-Fei. Effects of typical plant on soil microbial communities in an Inner Mongolia grassland [J]. Chin J Plant Ecol, 2014, 38(2): 201-208. |
| [9] | Hongzheng Ma, Shanshan Li, Song Ge, Silan Dai, Wenli Chen. Isolation of SSR markers for two related second-generation energy crop species, Miscanthus nepalensis and M. nudipes (Poaceae) [J]. Biodiv Sci, 2011, 19(5): 535-542. |
| [10] | Yaqiong Du, Zicheng Wang. Methylation-sensitive Amplified Polymorphism Analysis of DNA Methylation in Arabidopsis Under Mannitol Treatment [J]. Chinese Bulletin of Botany, 2011, 46(3): 285-292. |
| [11] | Ying Xu, Jing Xu, Jiyin Gao, Wenju Zhang. Polymorphism of the Internal Transcribed Spacer of rDNA in Camellia——an Escape from Concerted Evolution [J]. Chinese Bulletin of Botany, 2011, 46(2): 162-169. |
| [12] | ZHANG Yun-Hong, HOU Yan, LOU An-Ru. Population genetic diversity of Rhodiola dumulosa in Northern China inferred from AFLP makers [J]. Chin J Plant Ecol, 2010, 34(9): 1084-1094. |
| [13] | LIU Wei, WANG Xi, GAN You-Min, HUANG Lin-Kai, XIE Wen-Gang, MIAO Jia-Min. GENETIC DIVERSITY OF KOBRESIA PYGMAEA POPULATIONS ALONG A GRAZING GRADIENT [J]. Chin J Plant Ecol, 2009, 33(5): 966-973. |
| [14] | Lichuan Dai, Minglong Zhang, Jiye Liu, Xiaobai Li, Hairui Cui. Genetic diversity in Chinese rapeseed (Brassica napus) cultivars based on EST-SSR markers [J]. Biodiv Sci, 2009, 17(5): 482-489. |
| [15] | Longqian Xiao, Hua Zhu. Intra-genomic polymorphism in the internal transcribed spacer (ITS) regions of Cycas revoluta: evidence of incomplete concerted evolution [J]. Biodiv Sci, 2009, 17(5): 476-481. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||