INTRODUCTION Genetic diversity is the natural attribute of organisms formed in the long-term evolution process, which refers to the sum of all genetic variations of different individuals within a species or a group. Pepper (Capsicumspp.), a popular vegetable crop in China, is cultivated extensively with a substantial annual yield. Nevertheless, the widespread adoption of commercial pepper varieties has led to a gradual reduction in its genetic diversity, resulting in an increasing homogenization of germplasm resources. Given that germplasm forms the foundation for crop genetic improvement, maintaining rich genetic diversity is crucial for effective breeding.
RATIONALE On the other hand, morphological markers serve as a fundamental approach for investigating plant phenotypic diversity, as they allow researchers to evaluate the variations in marked samples through the observation of plant agronomic traits. On the other hand, molecular markers, particularly SSR (simple sequence repeat) markers, have gained widespread application in the breeding of new crop varieties and related fields due to their accuracy and reliability. This study focused on analyzing the diversity of 146 pepper germplasms in Inner Mongolia by employing both morphological traits and SSR markers.
RESULTS We assessed 34 morphological traits of 146 pepper germplasms and evaluated their genetic diversity using 22 pairs of SSR primers. Our analysis revealed a high degree of diversity in the traits of these pepper lines. The results of phenotypic trait diversity analysis showed that the coefficient of variation of quality traits and quantitative traits ranged from 8.22% to 267.58% and 14.35% to 72.51%, respectively, and the Shannon-Wiener diversity index ranged from 0.04 to 1.91 and 1.58 to 2.02, respectively. The genetic diversity of pepper germplasm resources was rich. A total of 102 alleles were detected by 22 pairs of SSR fluorescent molecular markers, with an average of 4.636 alleles per pair of primers. The effective allelic variation ranged from 1.191 to 5.311, the Shannon-Wiener diversity index ranged from 0.345 to 2.056, and the polymorphic information content (PIC) ranged from 0.153 to 0.795. The average genetic distance of 146 pepper germplasm resources was 0.429. Phenotypic value clustering and principal component analysis categorized the 146 accessions into six distinct groups, while the analysis of the SSR marker data divided them into seven groups. Population genetic structure analysis further delineated the 146 pepper germplasms into two main groups. Most of these germplasms were high-generation breeding lines with high homozygosity. However, gene introgression was observed within Group1 and Group2.
CONCLUSION This study is the systematic analysis of the morphological characteristics, genetic diversity and population structure of 146 pepper germplasms, particularly by utilizing SSR fluorescent molecular markers. The high diversity in both morphological traits and genetic markers of these pepper germplasm resources indicated significant genetic diffe- rences in them. Consequently, this study provides a better understanding of the germplasm diversity and population genetic structure of these 146 pepper germplasms, establishing a theoretical foundation for future variety breeding efforts.
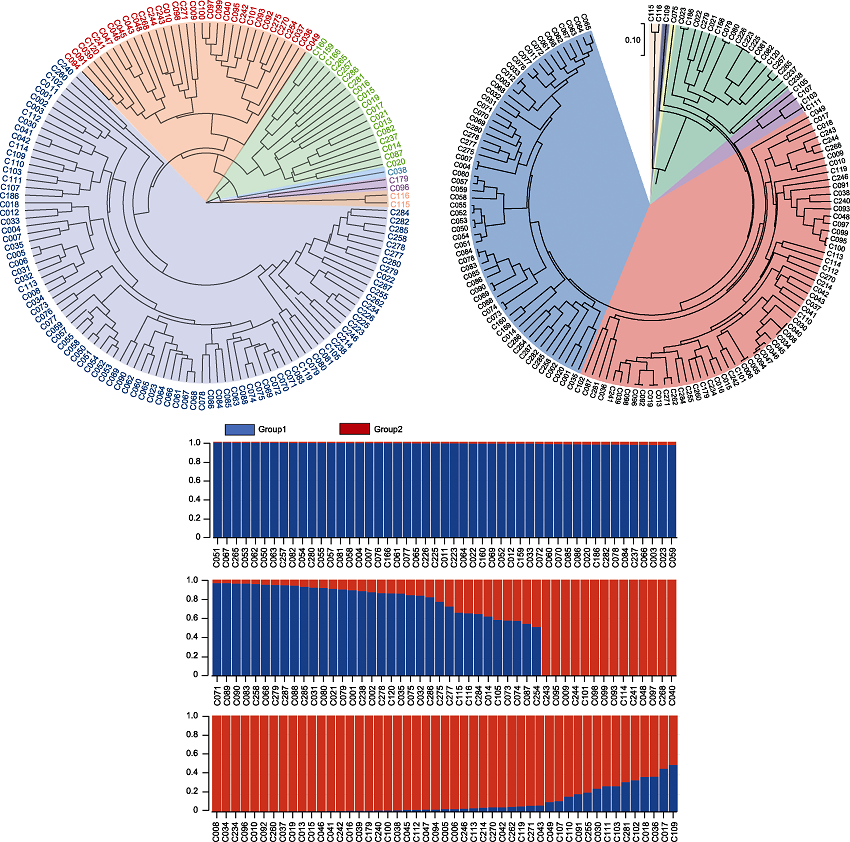
Genetic diversity analysis of 146 pepper germplasms. 146 pepper germplasms were categorized into six groups based on phenotypic markers and seven groups based on molecular markers, but the correlation between these clusters was weak (r=0.3967). Population genetic structure analysis further divided the germplasms into two distinct groups.

