

Chinese Bulletin of Botany ›› 2017, Vol. 52 ›› Issue (2): 179-187.DOI: 10.11983/CBB16041 cstr: 32102.14.CBB16041
Previous Articles Next Articles
Wang Qian, Sun Wenjing, Bao Ying*( )
)
Received:2016-03-08
Accepted:2016-08-08
Online:2017-03-01
Published:2017-04-05
Contact:
Bao Ying
About author:# Co-first authors
Wang Qian, Sun Wenjing, Bao Ying. Evolutionary Pattern of the GBSS Gene Family in Plants[J]. Chinese Bulletin of Botany, 2017, 52(2): 179-187.
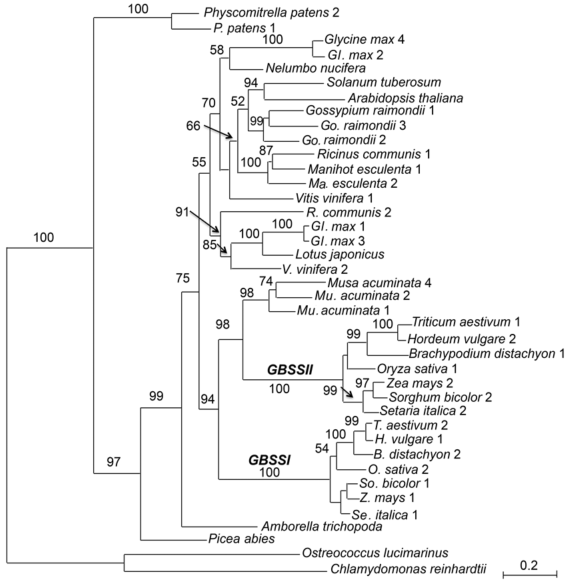
Figure 1 The maximum likelihood tree of the granule-bound starch synthase gene family based on amino acid sequences of 22 plant speciesNumbers above the branches indicate bootstrap values above 50%. Numbers following species names indicate different gene loci as listed in Table 1.
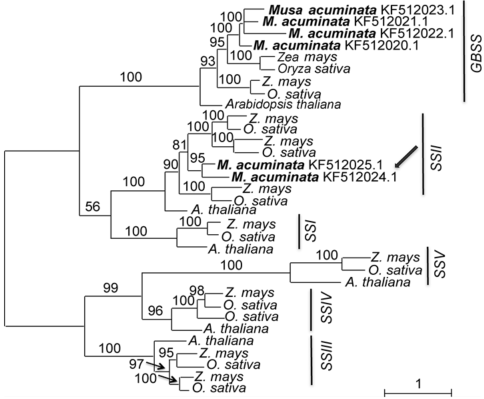
Figure 2 Maximum likelihood tree of 4 species based on amino acid homologous sequences of the starch synthase genesNumbers above the branches indicate bootstrap values above 50%. Two genes of Musa acuminata indicated by arrow should be SSII rather than GBSS.
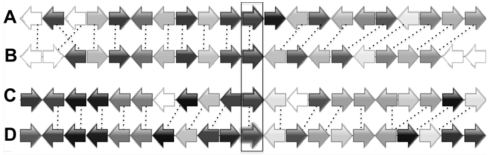
Figure 3 Synteny alignment of the chromosome regions with 4 GBSS homologous genes in Glycine max(A) The chromosome region includes locus GM10G31540; (B) The chromosome region includes locus GM20G36040; (C) The chromosome region includes locus GM16G02110; (D) The chromosome region includes locus GM07G05580. Arrows indicate the direction of genes’ transcription. Homologous genes are shown in same colors. Four GBSS genes are in the frame.
| [1] | 包颖, 杜家潇, 景翔, 徐思 (2015). 药用野生稻叶中淀粉合成酶基因家族的序列分化和特异表达. 植物学报50, 683-690. |
| [2] | Ahuja G, Jaiswal S, Hucl P, Chibbar RN (2014). Wheat genome specific granule-bound starch synthase I differentially influence grain starch synthesis.Carbohydr Polym 114, 87-94. |
| [3] | Alison MS (2012). Starch in the Arabidopsis plant.Starch 64, 421-434. |
| [4] | Ball S, Colleoni C, Cenci U, Raj JN, Tirtiaux C (2011). The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis.J Exp Bot 62, 1775-1801. |
| [5] | Baranov Iu O, Slishchuk HI, Volkova NE, SyvolapIu M (2014). Bioinformatic analysis of maize granule-bound starch synthase gene.Tsitol Genet 48, 18-23. |
| [6] | Criscuolo A (2011). morePhyML: improving the phylogenetic tree space exploration with PhyML 3.Mol Phylogenet Evol 61, 944-948. |
| [7] | Deschamps P, Moreau H, Worden AZ, Dauvillee D, Ball SG (2008). Early gene duplication within chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts.Genetics 178, 2373-2387. |
| [8] | Dian W, Jiang H, Chen Q, Liu F, Wu P (2003). Cloning and characterization of the granule-bound starch synthase II gene in rice: gene expression is regulated by the nitrogen level, sugar and circadian rhythm.Planta 218, 261-268. |
| [9] | Fasahat P, Rahman S, Ratnam W (2014). Genetic controls on starch amylose content in wheat and rice grains.J Genet 93, 279-292. |
| [10] | Fulton DC, Edwards A, Pilling E, Robinson HL, Fahy B, Seale R, Kato L, Donald AM, Geigenberger P, Martin C, Smith AM (2002). Role of granule-bound starch synthase in determination of amylopectin structure and starch granule morphology in potato.J Biol Chem 277, 10834-10841. |
| [11] | Guzman C, Alvarez JB (2015). Wheat waxy proteins: polymorphism, molecular characterization and effects on starch properties.Theor Appl Genet 9, 1049-1060. |
| [12] | Hirose T, Hashida Y, Aoki N, Okamura M, Yonekura M, Ohto C, Terao T, Ohsugi R (2014). Analysis of gene- disruption mutants of a sucrose phosphate synthase gene in rice,OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant Sci 225, 102-106. |
| [13] | Hoai TT, Matsusaka H, Toyosawa Y, Suu TD, Satoh H, Kumamaru T (2014). Influence of single-nucleotide poly- morphisms in the gene encoding granule-bound starch synthase I on amylose content in Vietnamese rice cultivars.Breed Sci 64, 142-148. |
| [14] | Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y (2010). Starch biosynthesis in cereal endosperm.Plant Physiol Biochem 48, 383-392. |
| [15] | Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007). Clustal W and Clustal X version 2.0.Bioinformatics 23, 2947-2948. |
| [16] | Miao H, Sun P, Liu W, Xu B, Jin Z (2014). Identification of genes encoding granule-bound starch synthase involved in amylose metabolism in banana fruit.PLoS One 9, e88077. |
| [17] | Ohdan T, Francisco PB, Sawada TJ, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot 56, 3229-3244. |
| [18] | Orzechowski S (2008). Starch metabolism in leaves.Acta Biochim Pol 55, 435-445. |
| [19] | Patron NJ, Smith AM, Fahy BF, Hylton CM, Naldrett MJ, Rossnagel BG, Denyer K (2002). The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5'-non-coding region.Plant Physiol 130, 190-198. |
| [20] | Tsai CY (1974). The function of the waxy locus in starch synthesis in maize endosperm.Biochem Genet 11, 83-96. |
| [21] | Vrinten PL, Nakamura T (2000). Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues.Plant Physiol 122, 255-264. |
| [22] | Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench.Bioinformatics 25, 1189-1191. |
| [23] | Yan HB, Pan XX, Jiang HW, Wu GJ (2009). Comparison of the starch synthesis genes between maize and rice: copies, chromosome location and expression divergence.Theor Appl Genet 119, 815-825. |
| [24] | Zhu L, Gu M, Meng X, Cheung SC, Yu H, Huang J, Sun Y, Shi Y, Liu Q (2012). High-amylose rice improves indices of animal health in normal and diabetic rats.Plant Biotechnol J 10, 353-362. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||