

Chinese Bulletin of Botany ›› 2015, Vol. 50 ›› Issue (2): 171-179.DOI: 10.3724/SP.J.1259.2015.00171 cstr: 32102.14.SP.J.1259.2015.00171
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Zili Wu1, 2, Mengyao Yu2, Lu Chen2, Jing Wei2, Xiaoqin Wang3, Yong Hu2, Yan Yan2, Ping Wan2, *
Received:2014-10-27
Accepted:2015-02-02
Online:2015-03-01
Published:2015-04-10
Contact:
Wan Ping
About author:? These authors contributed equally to this paper
Zili Wu, Mengyao Yu, Lu Chen, Jing Wei, Xiaoqin Wang, Yong Hu, Yan Yan, Ping Wan. Transcriptome Analysis of Physcomitrella patens Response to Cadmium Stress by Bayesian Network[J]. Chinese Bulletin of Botany, 2015, 50(2): 171-179.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.chinbullbotany.com/EN/10.3724/SP.J.1259.2015.00171
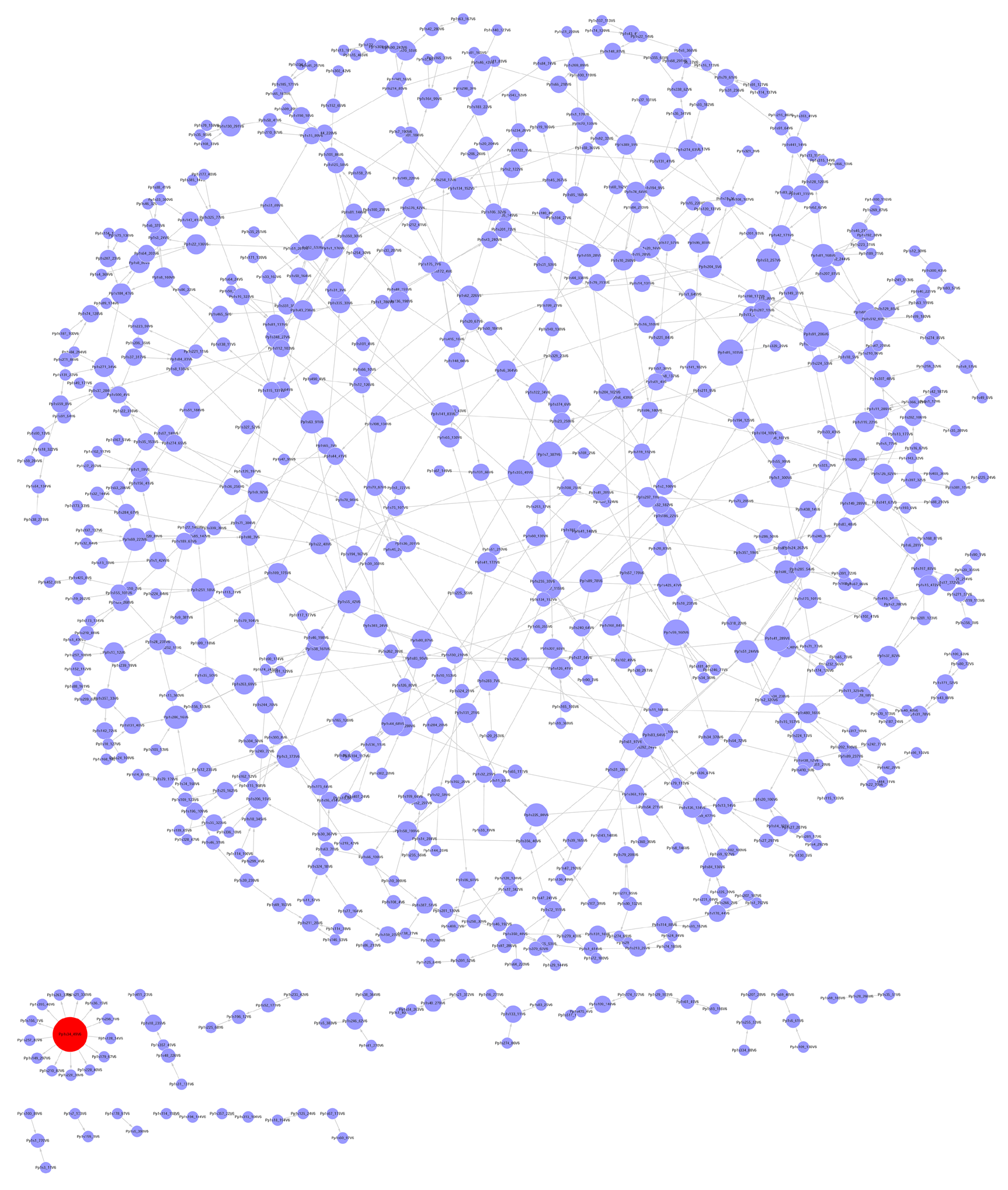
The gene regulatory network of Physcomitrella patens under cadmium stress The network consists of 704 nodes (genes) and 788 edges (regulation relation). The degree of the red node is thirteen
| Type | GO number | Biological processes | P value |
|---|---|---|---|
| 1 | GO:0007018 | Microtubule-based movement | 2.80E-06 |
| GO:0007017 | Microtubule-based process | 6.90E-05 | |
| GO:0007010 | Cytoskeleton organization | 0.018 8 | |
| 2 | GO:0006260 | DNA replication | 0.001 7 |
| GO:0006259 | DNA metabolic process | 0.002 5 | |
| GO:0006302 | Double-strand break repair | 0.015 3 | |
| GO:0000723 | Telomere maintenance | 0.033 4 | |
| GO:0032200 | Telomere organization | 0.033 4 | |
| 3 | GO:0007276 | Gamete generation | 0.033 4 |
| GO:0007292 | Female gamete generation | 0.033 4 | |
| GO:0019953 | Sexual reproduction | 0.033 4 | |
| 4 | GO:0019627 | Urea metabolic process | 0.033 4 |
| GO:0071941 | Nitrogen cycle metabolic process | 0.033 4 |
Table 1 The enriched GO terms in Physcomitrella patens gene regulation network under cadmium stress
| Type | GO number | Biological processes | P value |
|---|---|---|---|
| 1 | GO:0007018 | Microtubule-based movement | 2.80E-06 |
| GO:0007017 | Microtubule-based process | 6.90E-05 | |
| GO:0007010 | Cytoskeleton organization | 0.018 8 | |
| 2 | GO:0006260 | DNA replication | 0.001 7 |
| GO:0006259 | DNA metabolic process | 0.002 5 | |
| GO:0006302 | Double-strand break repair | 0.015 3 | |
| GO:0000723 | Telomere maintenance | 0.033 4 | |
| GO:0032200 | Telomere organization | 0.033 4 | |
| 3 | GO:0007276 | Gamete generation | 0.033 4 |
| GO:0007292 | Female gamete generation | 0.033 4 | |
| GO:0019953 | Sexual reproduction | 0.033 4 | |
| 4 | GO:0019627 | Urea metabolic process | 0.033 4 |
| GO:0071941 | Nitrogen cycle metabolic process | 0.033 4 |
| Gene identification number | The homologous gene in Arabidopsis thaliana | Function |
|---|---|---|
| Pp1s34_49V6 | AT2G27290.1 | Protein of unknown function (DUF1279) |
| Pp1s352_53V6 | AT3G57060.2 | Chromosome condensation |
| Pp1s59_160V6 | AT3G44750.1 | Histone deacetylase 3 |
| Pp1s57_179V6 | AT5G18140.1 | Chaperone DnaJ-domain superfamily protein |
| Pp1s204_5V6 | AT5G20935.1 | Protein of unknown function |
| Pp1s68_291V6 | AT5G57590.1 | Adenosylmethionine-8-amino-7-oxononanoate transaminases |
| Pp1s13_454V6 | AT5G54910.1 | DEA (D/H)-box RNA helicase family protein |
| Pp1s133_35V6.1 | AT1G08260.1 | DNA polymerase epsilon catalytic subunit |
| Pp1s259_32V6.1 | AT1G79690.1 | Dipeptidyl-peptidase activity, hydrolase activity |
| Pp1s175_94V6.1 | AT4G01130.1 | GDSL-like Lipase/Acylhydrolase superfamily protein involved in lipid metabolic process |
Table 2 A list of ten genes selected for qPCR analysis
| Gene identification number | The homologous gene in Arabidopsis thaliana | Function |
|---|---|---|
| Pp1s34_49V6 | AT2G27290.1 | Protein of unknown function (DUF1279) |
| Pp1s352_53V6 | AT3G57060.2 | Chromosome condensation |
| Pp1s59_160V6 | AT3G44750.1 | Histone deacetylase 3 |
| Pp1s57_179V6 | AT5G18140.1 | Chaperone DnaJ-domain superfamily protein |
| Pp1s204_5V6 | AT5G20935.1 | Protein of unknown function |
| Pp1s68_291V6 | AT5G57590.1 | Adenosylmethionine-8-amino-7-oxononanoate transaminases |
| Pp1s13_454V6 | AT5G54910.1 | DEA (D/H)-box RNA helicase family protein |
| Pp1s133_35V6.1 | AT1G08260.1 | DNA polymerase epsilon catalytic subunit |
| Pp1s259_32V6.1 | AT1G79690.1 | Dipeptidyl-peptidase activity, hydrolase activity |
| Pp1s175_94V6.1 | AT4G01130.1 | GDSL-like Lipase/Acylhydrolase superfamily protein involved in lipid metabolic process |
| 1 | Cabot C, Gallego B, Martos S, Barceló J, Poschenrieder C (2013). Signal cross talk in Arabidopsis exposed to cadmium, silicon, and Botrytis cinerea.Planta 237, 337-349. |
| 2 | Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, Shen YP, Zhang L, Deng XH, Luo JC, Deng XW, Chen ZL, Gu HY, Qu LJ (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family.Plant Mol Biol 60, 107-124. |
| 3 | Chmielowska-Bak J, Deckert J (2012). A common res- ponse to common danger? Comparison of animal and plant signaling pathways involved in cadmium sensing.J Cell Commun Signal 6, 191-204. |
| 4 | Chmielowska-Bak J, Deckert J (2013). Nitric oxide med- iates Cd-dependent induction of signaling-associated genes.Plant Signal Behav 8, e26664. |
| 5 | Chmielowska-Bak J, Gzyl J, Rucińska-Sobkowiak R, Arasimowicz-Jelonek M, Deckert J (2014). The new insights into cadmium sensing.Front Plant Sci 5, 245. |
| 6 | Corradi MG, Gorbi G, Ricci A, Torelli A, Bassi M (1995). Chromium-induced sexual reproduction gives rise to a Cr-tolerant progeny in Scenedesmus acutus.Ecotoxicol Environ Saf 32, 12-18. |
| 7 | DalCorso G, Farinati S, Furini A (2010). Regulatory networks of cadmium stress in plants.Plant Signal Behav 5, 663-667. |
| 8 | Dovgalyuk A, Kalynyak T, Blume YB (2003). Heavy metals have a different action from aluminium in disrupting microtubules in Allium cepa meristematic cells.Cell Biol Int 27, 193-195. |
| 9 | Ercal N, Gurer-Orhan H, Aykin-Burns N (2001). Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage.Curr Top Med Chem 1, 529-539. |
| 10 | Farinati S, DalCorso G, Varotto S, Furini A (2010). The Brassica juncea BjCdR15, an ortholog of Arabidopsis TGA3, is a regulator of cadmium uptake, transport and accumulation in shoots and confers cadmium tolerance in transgenic plants.New Phytol 185, 964-978. |
| 11 | Fojtová M, Fulnečková J, Fajkus J, Kovařík A (2002). Recovery of tobacco cells from cadmium stress is accompanied by DNA repair and increased telomerase activity.J Exp Bot 53, 2151-2158. |
| 12 | Hart JJ, Welch RM, Norvell WA, Sullivan LA, Kochian LV (1998). Characterization of cadmium binding, uptake, and translocation in intact seedlings of bread and durum wheat cultivars.Plant Physiol 116, 1413-1420. |
| 13 | Hartwig A, Schwerdtle T (2002). Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications.Toxicol Lett 127, 47-54. |
| 14 | Hepler PK, Hush JM (1996). Behavior of microtubules in living plant cells.Plant Physiol 112, 455-461. |
| 15 | Herbette S, Taconnat L, Hugouvieux V, Piette L, Magniette MLM, Cuine S, Auroy P, Richaud P, Forestier C, Bourguignon J, Renou JP, Vavasseur A, Leonhardt N (2006). Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88, 1751-1765. |
| 16 | Hsu YT, Kao CH (2003). Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings.Plant Cell Environ 26, 867-874. |
| 17 | Huang JJ, Okuka M, Lu WS, Tsibris JCM, McLean MP, Keefe DL, Liu L (2013). Telomere shortening and DNA damage of embryonic stem cells induced by cigarette smoke.Reprod Toxicol 35, 89-95. |
| 18 | Liu DH, Xue P, Meng QM, Zou J, Gu JG, Jiang WS (2009). Pb/Cu effects on the organization of microtubule cyto- skeleton in interphase and mitotic cells of Allium sativum L.Plant Cell Rep 28, 695-702. |
| 19 | Liu XM, Kim KE, Kim KC, Nguyen XC, Han HJ, Jung MS, Kim HS, Kim SH, Park HC, Yun DJ, Chung WS (2010). Cadmium activates Arabidopsis MPK3 and MPK6 via accumulation of reactive oxygen species.Phytochemistry 71, 614-618. |
| 20 | Ma WW, Xu WZ, Xu H, Chen YS, He ZY, Ma M (2010). Nitric oxide modulates cadmium influx during cadmium- induced programmed cell death in tobacco BY-2 cells.Planta 232, 325-335. |
| 21 | Nzengue Y, Steiman R, Garrel C, Lefèbvre E, Guiraud P (2008). Oxidative stress and DNA damage induced by cadmium in the human keratinocyte HaCaT cell line: role of glutathione in the resistance to cadmium.Toxicology 243, 193-206. |
| 22 | Oono Y, Yazawa T, Kawahara Y, Kanamori H, Kobayashi F, Sasaki H, Mori S, Wu J, Handa H, Itoh T, Matsumoto T (2014). Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice. PLoS One 9, e96946. |
| 23 | Přibyl P, Cepák V, Zachleder V (2008). Cytoskeletal alterations in interphase cells of the green alga Spirogyra decimina in response to heavy metals exposure: II. The effect of aluminium, nickel and copper.Toxicol In Vitro 22, 1160-1168. |
| 24 | Qi XT, Zhang YX, Chai TY (2007). Characterization of a novel plant promoter specifically induced by heavy metal and identification of the promoter regions conferring he- avy metal responsiveness.Plant Physiol 143, 50-59. |
| 25 | Rai V, Vajpayee P, Singh SN, Mehrotra S (2004). Effect of chromium accumulation on photosynthetic pigments, oxidative stress defense system, nitrate reduction, proline level and eugenol content of Ocimum tenuiflorum L.Plant Sci 167, 1159-1169. |
| 26 | Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin IT, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Cho SH, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van de Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants.Science 319, 64-69. |
| 27 | Roth U, von Roepenack-Lahaye E, Clemens S (2006). Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+.J Exp Bot 57, 4003-4013. |
| 28 | Rother M, Krauss GJ, Grass G, Wesenberg D (2006). Sulphate assimilation under Cd2+ stress in Physcomitrella patens—combined transcript, enzyme and metabolite profiling. Plant Cell Environ 29, 1801-1811. |
| 29 | Singh I, Shah K (2014). Evidences for structural basis of altered ascorbate peroxidase activity in cadmium- stressed rice plants exposed to jasmonate.Biometals 27, 247-263. |
| 30 | van de Mortel JE, Schat H, Moerland PD, Ver Loren van Themaat E, van der Ent S, Blankestijn H, Ghandilyan A, Tsiatsiani S, Aarts MG (2008). Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens.Plant Cell Environ 31, 301-324. |
| 31 | Wang YC, Gao CQ, Liang YN, Wang C, Yang CP, Liu GF (2010). A novel bZIP gene from Tamarix hispida mediates physiological responses to salt stress in tobacco plants.J Plant Physiol 167, 222-230. |
| 32 | Weber M, Trampczynska A, Clemens S (2006). Compara- tive transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd2+-hypertolerant facultative metallophyte Arabidopsis halleri.Plant Cell Environ 29, 950-963. |
| 33 | Xiong J, Fu G, Tao L, Zhu C (2010). Roles of nitric oxide in alleviating heavy metal toxicity in plants.Arch Biochem Biophys 497, 13-20. |
| 34 | Xiong J, Lu H, Lu KX, Duan YX, An LY, Zhu C (2009). Cadmium decreases crown root number by decreasing endogenous nitric oxide, which is indispensable for crown root primordia initiation in rice seedlings.Planta 230, 599-610. |
| 35 | Ye Y, Li Z, Xing D (2013). Nitric oxide promotes MPK6-mediated caspase-3-like activation in cadmium- induced Arabidopsis thaliana programmed cell death.Plant Cell Environ 36, 1-15. |
| 36 | Yourtchi MS, Bayat H (2013). Effect of cadmium toxicity on growth, cadmium accumulation and macronutrient content of durum wheat (Dena CV.).Int J Agri Crop Sci 6, 1099-1103. |
| [1] | Xiong Lianglin, Liang Guolu, Guo Qigao, Jing Danlong. Advances in the Regulation of Alternative Splicing of Genes in Plants in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2025, 60(3): 435-448. |
| [2] | Jing Xia, Yuchun Rao, Danyun Cao, Yi Wang, Linxin Liu, Yating Xu, Wangshu Mou, Dawei Xue. Research Progress on the Regulatory Mechanisms of OsACS and OsACO in Rice Ethylene Biosynthesis [J]. Chinese Bulletin of Botany, 2024, 59(2): 291-301. |
| [3] | Zhengming Luo, Jinxian Liu, Bianhua Zhang, Yanying Zhou, Aihua Hao, Kai Yang, Baofeng Chai. Diversity characteristics and driving factors of soil protist communities in subalpine meadow at different degradation stages [J]. Biodiv Sci, 2023, 31(8): 23136-. |
| [4] | ZHANG Zhong-Fu, WANG Si-Hai, YANG Wei, CHEN Jian. Response of rhizosphere microbial community structure and functional characteristics to health status of Malania oleifera [J]. Chin J Plant Ecol, 2023, 47(7): 1020-1031. |
| [5] | Yinger Mao, Xiumei Zhou, Nan Wang, Xiuxiu Li, Yuke You, Shangbin Bai. Impact of Phyllostachys edulis expansion to Chinese fir forest on the soil bacterial community [J]. Biodiv Sci, 2023, 31(6): 22659-. |
| [6] | Cai Shuyu, Liu Jianxin, Wang Guofu, Wu Liyuan, Song Jiangping. Regulatory Mechanism of Melatonin on Tomato Seed Germination Under Cd2+ Stress [J]. Chinese Bulletin of Botany, 2023, 58(5): 720-732. |
| [7] | Dai Ruohui, Qian Xinyu, Sun Jinglei, Lu Tao, Jia Qiwei, Lu Tianqi, Lu Mei, Rao Yuchun. Research Progress on the Mechanisms of Leaf Color Regulation and Related Genes in Rice [J]. Chinese Bulletin of Botany, 2023, 58(5): 799-812. |
| [8] | BAI Xue, LI Yu-Jing, JING Xiu-Qing, ZHAO Xiao-Dong, CHANG Sha-Sha, JING Tao-Yu, LIU Jin-Ru, ZHAO Peng-Yu. Response mechanisms of millet and its rhizosphere soil microbial communities to chromium stress [J]. Chin J Plant Ecol, 2023, 47(3): 418-433. |
| [9] | Wen Zhao, Dandan Wang, Mumin Reyila, Kaichuan Huang, Shun Liu, Baokai Cui. Soil microbial community structure of Larix gmelinii forest in the Aershan area [J]. Biodiv Sci, 2023, 31(2): 22258-. |
| [10] | Fan Xia, Jing Yang, Jian Li, Yang Shi, Lixin Gai, Wenhua Huang, Jingwei Zhang, Nan Yang, Fuli Gao, Yingying Han, Weidong Bao. Gut bacterial composition of four leopard cat subpopulations in Beijing [J]. Biodiv Sci, 2022, 30(9): 22103-. |
| [11] | Li Cong, Qi Lijuan, Gu Xiaofeng, Li Jigang. Research Progress on TZP, a Novel Key Regulator of Light Signal Transduction in Plants [J]. Chinese Bulletin of Botany, 2022, 57(5): 579-587. |
| [12] | Haixia Xu, Jing He, Hang Yi, Li Wang. Sex Specific Response Mechanism of Transcriptome in Both Male and Female Marchantia polymorpha Under Cadmium Stress [J]. Chinese Bulletin of Botany, 2022, 57(2): 182-196. |
| [13] | Yixin Sun, Yingbin Li, Yuhui Li, Bing Li, Xiaofang Du, Qi Li. Application of high-throughput sequencing technique in the study of nematode diversity [J]. Biodiv Sci, 2022, 30(12): 22266-. |
| [14] | Cheng Gao, Liang-Dong Guo. Progress on microbial species diversity, community assembly and functional traits [J]. Biodiv Sci, 2022, 30(10): 22429-. |
| [15] | Qifeng Lu, Zhihuan Huang, Wenhua Luo. Characterization of complete chloroplast genome in Firmiana kwangsiensis and F. danxiaensis with extremely small populations [J]. Biodiv Sci, 2021, 29(5): 586-595. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||