

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (6): 829-839.DOI: 10.11983/CBB18003 cstr: 32102.14.CBB18003
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Zhang Deng, Li Jingjian, Zhang Mengjie, Bao Yutao, Yang Xiao, Xu Wuyun, Ouyang Kunxi, Chen Xiaoyang*( )
)
Received:2018-01-03
Online:2018-11-01
Published:2018-12-05
Contact:
Chen Xiaoyang
Zhang Deng, Li Jingjian, Zhang Mengjie, Bao Yutao, Yang Xiao, Xu Wuyun, Ouyang Kunxi, Chen Xiaoyang. Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis in Neolamarckia cadamba[J]. Chinese Bulletin of Botany, 2018, 53(6): 829-839.
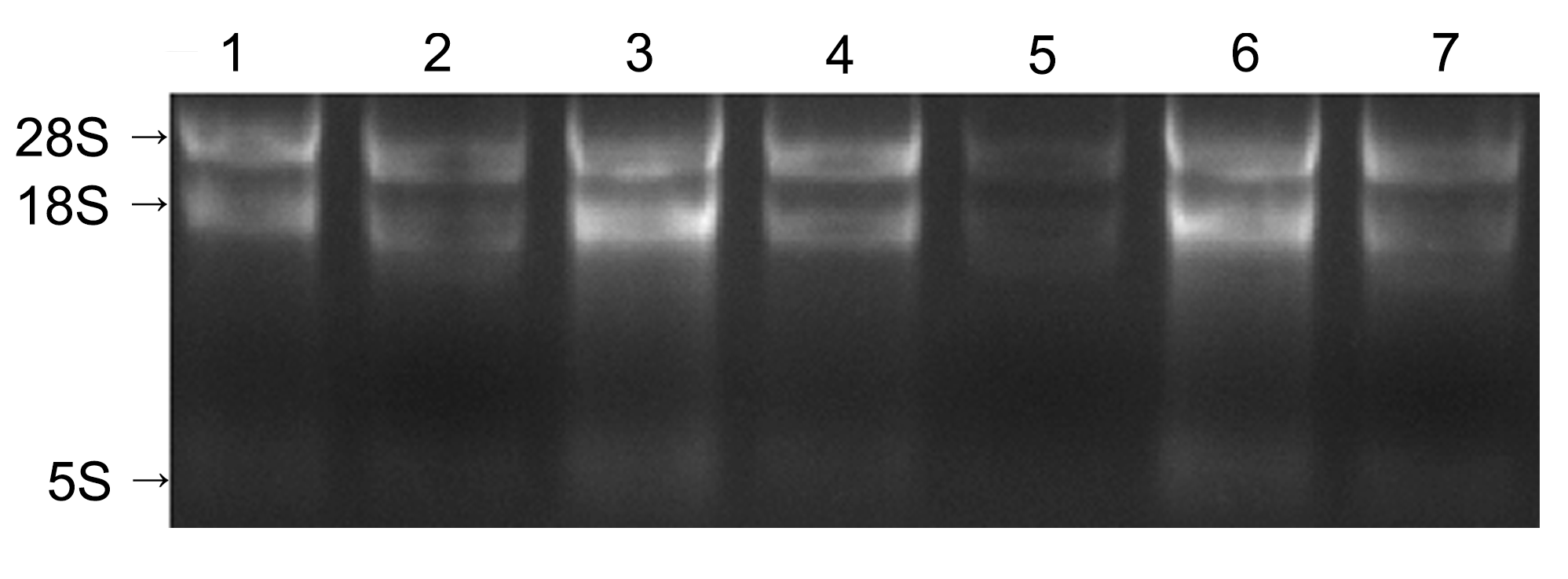
Figure 1 Electrophoresis of total RNA extracted from different tissues of Neolamarckia cadamba1: Bark; 2: Flower; 3: Cambium; 4: Leaf; 5: Root; 6: Bud; 7: Fruit
| Tissues | A260/A280 | A260/A230 | Yield (ng?μL-1) |
|---|---|---|---|
| Root | 2.19 | 2.1 | 107.5 |
| Bud | 2.12 | 2.14 | 410.6 |
| Leaf | 2.13 | 1.96 | 258.5 |
| Flower | 2.19 | 2.22 | 114.8 |
| Fruit | 2.11 | 1.84 | 156.8 |
| Bark | 2.17 | 1.89 | 299.7 |
| Cambium | 2.14 | 2.11 | 331.8 |
Table 1 The quality of total RNA extracted from different tissues of Neolamarckia cadamba
| Tissues | A260/A280 | A260/A230 | Yield (ng?μL-1) |
|---|---|---|---|
| Root | 2.19 | 2.1 | 107.5 |
| Bud | 2.12 | 2.14 | 410.6 |
| Leaf | 2.13 | 1.96 | 258.5 |
| Flower | 2.19 | 2.22 | 114.8 |
| Fruit | 2.11 | 1.84 | 156.8 |
| Bark | 2.17 | 1.89 | 299.7 |
| Cambium | 2.14 | 2.11 | 331.8 |
| Name | UniGene ID | Reference gene (Rg) ID | F primer (5'-3') R primer (5'-3') | Amplicator length (bp) |
|---|---|---|---|---|
| Actin | comp52737_c0 | g1 | TGTAGTGGATGAATGCTTCTGTTAT | 95 |
| CTTCCTCCTACCAACTTCAAATG | ||||
| comp79635_c0 | g2 | CTTCTGAGGTTATGGAGCAATCT | 101 | |
| CGATAAATCAAAACTTCAAGCC | ||||
| Clathrin adaptor complexes medium | comp48976_c0 | g3 | CTCAGAGAACGCTGCTGACTAC | 161 |
| GAGCCAAGGGAAACAAGATAA | ||||
| Cyclophilin | comp67418_c0 | g4 | GGGGTCTCACGCTCTTTACT | 83 |
| GGATTGGATTGGGTTGGTT | ||||
| comp75463_c0 | g5 | CCCCAGCAAGAAGACCACT | 213 | |
| TTGACCATGAATCCCAACCA | ||||
| comp77969_c0 | g6 | ATAGCATCCCAACCGAACA | 187 | |
| CCCTCTTGCCTCCTGTGTAT | ||||
| Elongation factor 1α | comp87079_c1 | g7 | ACCAGCATCACCGTTCTTCA | 123 |
| GTCCTCGATTGCCACACCT | ||||
| comp87526_c0 | g8 | AATCAGACAGAAACCCCTCAA | 245 | |
| GAACCTCTCAATCACACGCTT | ||||
| Eukaryotic initiation factor | comp6386_c0 | g9 | GTTGAAACTTCTTGGACATCG | 250 |
| CTTGAGACACTGATTTGTATGAGA | ||||
| Farnesyl pyrophosphate synthase 1 | comp72548_c0 | g10 | TGATAATCTGGCTTCCACCTT | 112 |
| TGGGAGGAACTCAATCTCCTAC | ||||
| comp75377_c0 | g11 | TATCAGGCTCAGCATTCCACT | 212 | |
| TTGCCACAATAACACATCCAT | ||||
| F-boxkelch-repeat protein | comp78454_c0 | g12 | AAGGCCAATTCTGTTCAAGC | 143 |
| CCTAGAGGGAAAGACATGACTG | ||||
| comp78817_c0 | g13 | GCAAACGGGGTAAAAGGA | 102 | |
| AAAGGGTAAGAGTGACGACAGC | ||||
| GAPDH | comp78593_c0 | g14 | TGTTCCAAGTGGGCATTTAC | 247 |
| CGCTCTGAGGTGTTAATAAGTG | ||||
| comp80828_c1 | g15 | CTGAGCATTTTTTAGGCTTGTC | 151 | |
| TCAGATTCATGTGGCAGTCG | ||||
| GTP-binding nuclear protein | comp85262_c0 | g16 | TCTCGCAACCTGCCTCTT | 257 |
| TATCACTCCCATCTTCGCAC | ||||
| Phosphoenolpyruvate carboxylase-related kinase 1 | comp75525_c0 | g17 | CGACCTCACATTCCTCATTAC | 291 |
| ACATAGACCATCCAGAGCCCA | ||||
| comp80613_c0 | g18 | TACATAGACCATCCAGAGCCA | 112 | |
| GCAAAAGGGCAAGCAACAG | ||||
| Protein phosphatase 2A | comp81334_c1 | g19 | GGGCTTTCCATCCCATACC | 128 |
| AGCCTTAGGGGGATTGGAA | ||||
| comp52412_c0 | g20 | ATGTTGGATGATATTAGTGGTGTG | 161 | |
| TCATAGGAAAATAGACCTCTGGTT | ||||
| Ribosomal protein L | comp46755_c0 | g21 | CTGAGGATTGTTAGCAGTTGAC | 119 |
| ACCAGAAAACAGACCACCTAAG | ||||
| comp52434_c0 | g22 | AAGGAAGGTAAAGCAGGGAA | 177 | |
| GCATGGGCAGGGATATAAAC | ||||
| comp87976_c0 | g23 | CACGCAGCATAGCCAAAC | 157 | |
| AGGCAGTTCTCTGATTCTTTTG | ||||
| Name | UniGene ID | Reference gene (Rg) ID | F primer (5'-3') R primer (5'-3') | Amplicator length (bp) |
| Ribosomal protein S | comp65909_c1 | g24 | GCTATGGTAGTCTCCCGAAAG | 182 |
| GGGGGAACAAGACTAAGGGT | ||||
| comp67276_c0 | g25 | TTTTGTTTCCCCTCTTTGC | 97 | |
| AACCTTGAACAACCTGTGTAGAA | ||||
| comp71526_c0 | g26 | CGGTTACACAAGGTTGAATGA | 117 | |
| AGAGGGTCTGGATTTGAGTGA | ||||
| Ribulose 1,5-bisphosphate carboxylase | comp47386_c0 | g27 | CAGCACCGTAATCCATAAAAC | 226 |
| CAAGCAGCCCAGCAAGTC | ||||
| comp88001_c0 | g28 | ACAGGATGGGTAGAAAGAGGC | 210 | |
| AGGATTGAGCCGAATACAACG | ||||
| S-adenosylmethionine decarboxylase | comp44802_c0 | g29 | TCTTCGTGGCACTTCTCTCC | 133 |
| ACAGGGTGTTGACTTGTTTCC | ||||
| comp71874_c0 | g30 | ATAAGGTCTCTTCTTGTTCGTGTAG | 178 | |
| GACTGAACAGCAACAGGAATAAT | ||||
| comp80075_c0 | g31 | GCTGCCTGTGGGTCTCCTA | 85 | |
| GTAAACCCCAATGCTACTCCT | ||||
| Translation elongation factor | comp65909_c1 | g32 | GCTATGGTAGTCTCCCGAAAG | 184 |
| CTGGGGGAACAAGACTAAGG | ||||
| comp70791_c0 | g33 | TCAACCAACCGTTCCTACC | 195 | |
| ACAACAGTCCTTTGCCACC | ||||
| Tubulin α | comp70323_c2 | g34 | GGTGGTGGAACTGGCTCTG | 217 |
| GGCAAATGTCATAGATGGCTT | ||||
| comp76448_c4 | g35 | AAGGAGGGAATGAGTGGAG | 107 | |
| ACTATGGCAAGAAGTCAAAGC | ||||
| Tubulin B | comp66056_c0 | g36 | GCAAGAAAGCCTTCCTCCTAA | 153 |
| TTCCCAACAATGTCAAATCAA | ||||
| comp79707_c1 | g37 | TTCAGGAGAGTCAGCGAGC | 187 | |
| CATCGTCTTCATATTCCCCTT | ||||
| Ubiquitin conjugating enzyme | comp79182_c1 | g38 | TCCTTGCTTGTGGCGTCA | 213 |
| CACGGGTGTCAAATCTGGC | ||||
| Ubiquitin | comp67366_c0 | g39 | GACGGGAGGACCTTAGCA | 298 |
| CTCGGAGACGGAGAACAA | ||||
| comp82561_c0 | g40 | GCATTTGTGTCTTGCCTCTTTAT | 186 | |
| GCGATGAGCAACATTCCTTTA | ||||
| comp68357_c0 | g41 | TTTTTCAGCAAAGAACAACCG | 135 | |
| TGAAGACCCTCACTGGAAAGA | ||||
| Ubiquitin-protein ligase | comp87122_c0 | g42 | GGTTGGTGGTAGAGTTGTGACTC | 182 |
| CGAGCACTACCACGACACG | ||||
| comp87211_c0 | g43 | GCCCCTCCGTTAAACTCG | 122 | |
| GCCATACTCCCACCGAAAT | ||||
Table 2 Primer sequences of the candidate reference genes
| Name | UniGene ID | Reference gene (Rg) ID | F primer (5'-3') R primer (5'-3') | Amplicator length (bp) |
|---|---|---|---|---|
| Actin | comp52737_c0 | g1 | TGTAGTGGATGAATGCTTCTGTTAT | 95 |
| CTTCCTCCTACCAACTTCAAATG | ||||
| comp79635_c0 | g2 | CTTCTGAGGTTATGGAGCAATCT | 101 | |
| CGATAAATCAAAACTTCAAGCC | ||||
| Clathrin adaptor complexes medium | comp48976_c0 | g3 | CTCAGAGAACGCTGCTGACTAC | 161 |
| GAGCCAAGGGAAACAAGATAA | ||||
| Cyclophilin | comp67418_c0 | g4 | GGGGTCTCACGCTCTTTACT | 83 |
| GGATTGGATTGGGTTGGTT | ||||
| comp75463_c0 | g5 | CCCCAGCAAGAAGACCACT | 213 | |
| TTGACCATGAATCCCAACCA | ||||
| comp77969_c0 | g6 | ATAGCATCCCAACCGAACA | 187 | |
| CCCTCTTGCCTCCTGTGTAT | ||||
| Elongation factor 1α | comp87079_c1 | g7 | ACCAGCATCACCGTTCTTCA | 123 |
| GTCCTCGATTGCCACACCT | ||||
| comp87526_c0 | g8 | AATCAGACAGAAACCCCTCAA | 245 | |
| GAACCTCTCAATCACACGCTT | ||||
| Eukaryotic initiation factor | comp6386_c0 | g9 | GTTGAAACTTCTTGGACATCG | 250 |
| CTTGAGACACTGATTTGTATGAGA | ||||
| Farnesyl pyrophosphate synthase 1 | comp72548_c0 | g10 | TGATAATCTGGCTTCCACCTT | 112 |
| TGGGAGGAACTCAATCTCCTAC | ||||
| comp75377_c0 | g11 | TATCAGGCTCAGCATTCCACT | 212 | |
| TTGCCACAATAACACATCCAT | ||||
| F-boxkelch-repeat protein | comp78454_c0 | g12 | AAGGCCAATTCTGTTCAAGC | 143 |
| CCTAGAGGGAAAGACATGACTG | ||||
| comp78817_c0 | g13 | GCAAACGGGGTAAAAGGA | 102 | |
| AAAGGGTAAGAGTGACGACAGC | ||||
| GAPDH | comp78593_c0 | g14 | TGTTCCAAGTGGGCATTTAC | 247 |
| CGCTCTGAGGTGTTAATAAGTG | ||||
| comp80828_c1 | g15 | CTGAGCATTTTTTAGGCTTGTC | 151 | |
| TCAGATTCATGTGGCAGTCG | ||||
| GTP-binding nuclear protein | comp85262_c0 | g16 | TCTCGCAACCTGCCTCTT | 257 |
| TATCACTCCCATCTTCGCAC | ||||
| Phosphoenolpyruvate carboxylase-related kinase 1 | comp75525_c0 | g17 | CGACCTCACATTCCTCATTAC | 291 |
| ACATAGACCATCCAGAGCCCA | ||||
| comp80613_c0 | g18 | TACATAGACCATCCAGAGCCA | 112 | |
| GCAAAAGGGCAAGCAACAG | ||||
| Protein phosphatase 2A | comp81334_c1 | g19 | GGGCTTTCCATCCCATACC | 128 |
| AGCCTTAGGGGGATTGGAA | ||||
| comp52412_c0 | g20 | ATGTTGGATGATATTAGTGGTGTG | 161 | |
| TCATAGGAAAATAGACCTCTGGTT | ||||
| Ribosomal protein L | comp46755_c0 | g21 | CTGAGGATTGTTAGCAGTTGAC | 119 |
| ACCAGAAAACAGACCACCTAAG | ||||
| comp52434_c0 | g22 | AAGGAAGGTAAAGCAGGGAA | 177 | |
| GCATGGGCAGGGATATAAAC | ||||
| comp87976_c0 | g23 | CACGCAGCATAGCCAAAC | 157 | |
| AGGCAGTTCTCTGATTCTTTTG | ||||
| Name | UniGene ID | Reference gene (Rg) ID | F primer (5'-3') R primer (5'-3') | Amplicator length (bp) |
| Ribosomal protein S | comp65909_c1 | g24 | GCTATGGTAGTCTCCCGAAAG | 182 |
| GGGGGAACAAGACTAAGGGT | ||||
| comp67276_c0 | g25 | TTTTGTTTCCCCTCTTTGC | 97 | |
| AACCTTGAACAACCTGTGTAGAA | ||||
| comp71526_c0 | g26 | CGGTTACACAAGGTTGAATGA | 117 | |
| AGAGGGTCTGGATTTGAGTGA | ||||
| Ribulose 1,5-bisphosphate carboxylase | comp47386_c0 | g27 | CAGCACCGTAATCCATAAAAC | 226 |
| CAAGCAGCCCAGCAAGTC | ||||
| comp88001_c0 | g28 | ACAGGATGGGTAGAAAGAGGC | 210 | |
| AGGATTGAGCCGAATACAACG | ||||
| S-adenosylmethionine decarboxylase | comp44802_c0 | g29 | TCTTCGTGGCACTTCTCTCC | 133 |
| ACAGGGTGTTGACTTGTTTCC | ||||
| comp71874_c0 | g30 | ATAAGGTCTCTTCTTGTTCGTGTAG | 178 | |
| GACTGAACAGCAACAGGAATAAT | ||||
| comp80075_c0 | g31 | GCTGCCTGTGGGTCTCCTA | 85 | |
| GTAAACCCCAATGCTACTCCT | ||||
| Translation elongation factor | comp65909_c1 | g32 | GCTATGGTAGTCTCCCGAAAG | 184 |
| CTGGGGGAACAAGACTAAGG | ||||
| comp70791_c0 | g33 | TCAACCAACCGTTCCTACC | 195 | |
| ACAACAGTCCTTTGCCACC | ||||
| Tubulin α | comp70323_c2 | g34 | GGTGGTGGAACTGGCTCTG | 217 |
| GGCAAATGTCATAGATGGCTT | ||||
| comp76448_c4 | g35 | AAGGAGGGAATGAGTGGAG | 107 | |
| ACTATGGCAAGAAGTCAAAGC | ||||
| Tubulin B | comp66056_c0 | g36 | GCAAGAAAGCCTTCCTCCTAA | 153 |
| TTCCCAACAATGTCAAATCAA | ||||
| comp79707_c1 | g37 | TTCAGGAGAGTCAGCGAGC | 187 | |
| CATCGTCTTCATATTCCCCTT | ||||
| Ubiquitin conjugating enzyme | comp79182_c1 | g38 | TCCTTGCTTGTGGCGTCA | 213 |
| CACGGGTGTCAAATCTGGC | ||||
| Ubiquitin | comp67366_c0 | g39 | GACGGGAGGACCTTAGCA | 298 |
| CTCGGAGACGGAGAACAA | ||||
| comp82561_c0 | g40 | GCATTTGTGTCTTGCCTCTTTAT | 186 | |
| GCGATGAGCAACATTCCTTTA | ||||
| comp68357_c0 | g41 | TTTTTCAGCAAAGAACAACCG | 135 | |
| TGAAGACCCTCACTGGAAAGA | ||||
| Ubiquitin-protein ligase | comp87122_c0 | g42 | GGTTGGTGGTAGAGTTGTGACTC | 182 |
| CGAGCACTACCACGACACG | ||||
| comp87211_c0 | g43 | GCCCCTCCGTTAAACTCG | 122 | |
| GCCATACTCCCACCGAAAT | ||||
| Rg ID | M | Rg ID | M | Rg ID | M | Rg ID | M | Rg ID | M |
|---|---|---|---|---|---|---|---|---|---|
| g25 | 0.443 | g23 | 1.142 | g36 | 1.649 | g6 | 2.045 | g40 | 2.365 |
| g42 | 0.443 | g31 | 1.183 | g34 | 1.693 | g4 | 2.080 | g17 | 2.412 |
| g21 | 0.471 | g24 | 1.229 | g28 | 1.737 | g2 | 2.113 | g16 | 2.458 |
| g33 | 0.565 | g13 | 1.287 | g7 | 1.789 | g32 | 2.148 | g38 | 2.503 |
| g15 | 0.704 | g22 | 1.348 | g12 | 1.836 | g39 | 2.184 | g5 | 2.566 |
| g43 | 0.862 | g14 | 1.404 | g10 | 1.881 | g19 | 2.217 | g27 | 2.652 |
| g29 | 0.966 | g26 | 1.467 | g11 | 1.927 | g37 | 2.251 | g41 | 2.859 |
| g18 | 1.014 | g9 | 1.533 | g30 | 1.967 | g1 | 2.285 | ||
| g8 | 1.090 | g35 | 1.597 | g3 | 2.007 | g20 | 2.321 |
Table 3 Analysis of expression stability of reference genes by geNorm
| Rg ID | M | Rg ID | M | Rg ID | M | Rg ID | M | Rg ID | M |
|---|---|---|---|---|---|---|---|---|---|
| g25 | 0.443 | g23 | 1.142 | g36 | 1.649 | g6 | 2.045 | g40 | 2.365 |
| g42 | 0.443 | g31 | 1.183 | g34 | 1.693 | g4 | 2.080 | g17 | 2.412 |
| g21 | 0.471 | g24 | 1.229 | g28 | 1.737 | g2 | 2.113 | g16 | 2.458 |
| g33 | 0.565 | g13 | 1.287 | g7 | 1.789 | g32 | 2.148 | g38 | 2.503 |
| g15 | 0.704 | g22 | 1.348 | g12 | 1.836 | g39 | 2.184 | g5 | 2.566 |
| g43 | 0.862 | g14 | 1.404 | g10 | 1.881 | g19 | 2.217 | g27 | 2.652 |
| g29 | 0.966 | g26 | 1.467 | g11 | 1.927 | g37 | 2.251 | g41 | 2.859 |
| g18 | 1.014 | g9 | 1.533 | g30 | 1.967 | g1 | 2.285 | ||
| g8 | 1.090 | g35 | 1.597 | g3 | 2.007 | g20 | 2.321 |
| Rg ID | Stability value | Rg ID | Stability value | Rg ID | Stability value | Rg ID | Stability value | Rg ID | Stability value |
|---|---|---|---|---|---|---|---|---|---|
| g42 | 0.223 | g8 | 0.787 | g28 | 1.249 | g6 | 1.441 | g40 | 1.937 |
| g15 | 0.298 | g23 | 0.871 | g35 | 1.264 | g4 | 1.460 | g17 | 2.023 |
| g25 | 0.343 | g31 | 0.951 | g10 | 1.316 | g19 | 1.496 | g38 | 2.062 |
| g43 | 0.505 | g9 | 1.007 | g12 | 1.354 | g2 | 1.518 | g16 | 2.099 |
| g21 | 0.517 | g13 | 1.021 | g36 | 1.371 | g39 | 1.545 | g5 | 2.350 |
| g33 | 0.554 | g22 | 1.108 | g3 | 1.372 | g32 | 1.574 | g27 | 2.750 |
| g18 | 0.567 | g14 | 1.136 | g34 | 1.384 | g1 | 1.657 | g41 | 4.759 |
| g29 | 0.590 | g7 | 1.141 | g11 | 1.397 | g37 | 1.709 | ||
| g24 | 0.724 | g26 | 1.160 | g30 | 1.411 | g20 | 1.733 |
Table 4 Stability analysis of reference gene expression by NormFinder
| Rg ID | Stability value | Rg ID | Stability value | Rg ID | Stability value | Rg ID | Stability value | Rg ID | Stability value |
|---|---|---|---|---|---|---|---|---|---|
| g42 | 0.223 | g8 | 0.787 | g28 | 1.249 | g6 | 1.441 | g40 | 1.937 |
| g15 | 0.298 | g23 | 0.871 | g35 | 1.264 | g4 | 1.460 | g17 | 2.023 |
| g25 | 0.343 | g31 | 0.951 | g10 | 1.316 | g19 | 1.496 | g38 | 2.062 |
| g43 | 0.505 | g9 | 1.007 | g12 | 1.354 | g2 | 1.518 | g16 | 2.099 |
| g21 | 0.517 | g13 | 1.021 | g36 | 1.371 | g39 | 1.545 | g5 | 2.350 |
| g33 | 0.554 | g22 | 1.108 | g3 | 1.372 | g32 | 1.574 | g27 | 2.750 |
| g18 | 0.567 | g14 | 1.136 | g34 | 1.384 | g1 | 1.657 | g41 | 4.759 |
| g29 | 0.590 | g7 | 1.141 | g11 | 1.397 | g37 | 1.709 | ||
| g24 | 0.724 | g26 | 1.160 | g30 | 1.411 | g20 | 1.733 |
| Parameter | Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| g42 | g15 | g25 | g43 | g21 | g33 | g18 | g8 | g29 | |
| SD±CP | 0.513 | 0.736 | 0.840 | 0.966 | 0.692 | 0.610 | 1.153 | 1.477 | 1.280 |
| CV (% CP) | 1.804 | 3.530 | 3.522 | 3.745 | 3.008 | 2.091 | 5.015 | 6.600 | 5.474 |
| coeff. of corr. (r) | 0.851 | 0.871 | 0.833 | 0.737 | 0.297 | 0.520 | 0.846 | 0.966 | 0.849 |
| p-value | 0.015 | 0.011 | 0.020 | 0.059 | 0.515 | 0.232 | 0.016 | 0.001 | 0.016 |
Table 5 Stability analysis of reference gene expression by BestKeeper
| Parameter | Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| g42 | g15 | g25 | g43 | g21 | g33 | g18 | g8 | g29 | |
| SD±CP | 0.513 | 0.736 | 0.840 | 0.966 | 0.692 | 0.610 | 1.153 | 1.477 | 1.280 |
| CV (% CP) | 1.804 | 3.530 | 3.522 | 3.745 | 3.008 | 2.091 | 5.015 | 6.600 | 5.474 |
| coeff. of corr. (r) | 0.851 | 0.871 | 0.833 | 0.737 | 0.297 | 0.520 | 0.846 | 0.966 | 0.849 |
| p-value | 0.015 | 0.011 | 0.020 | 0.059 | 0.515 | 0.232 | 0.016 | 0.001 | 0.016 |
| Genes | G | N | B | O |
|---|---|---|---|---|
| g42 | 1 | 1 | 1 | 1 |
| g25 | 1 | 3 | 5 | 2 |
| g21 | 3 | 5 | 3 | 3 |
| g33 | 4 | 6 | 2 | 4 |
| g15 | 5 | 2 | 4 | 3 |
| g43 | 6 | 4 | 6 | 5 |
Table 6 The rank of total score by 3 analysis softwares
| Genes | G | N | B | O |
|---|---|---|---|---|
| g42 | 1 | 1 | 1 | 1 |
| g25 | 1 | 3 | 5 | 2 |
| g21 | 3 | 5 | 3 | 3 |
| g33 | 4 | 6 | 2 | 4 |
| g15 | 5 | 2 | 4 | 3 |
| g43 | 6 | 4 | 6 | 5 |
| [1] |
邓小梅, 欧阳昆唏, 张倩, 黄浩, 陈晓阳 (2011). 团花研究现状及发展思考. 中南林业科技大学学报 31, 90-95.
DOI URL |
| [2] |
姜琼, 王幼宁, 王利祥, 孙政玺, 李霞 (2015). 盐胁迫下大豆根组织定量PCR分析中内参基因的选择. 植物学报 50, 754-764.
DOI URL |
| [3] | 欧阳昆唏, 李俊成, 黄浩, 刘明骞,陈晓阳 (2013). 团花树α-扩展蛋白基因的克隆及表达分析. 林业科学 49, 62-71. |
| [4] | 孙美莲, 王云生, 杨冬青, 韦朝领, 高丽萍, 夏涛, 单育, 骆洋 (2010). 茶树实时荧光定量PCR分析中内参基因的选择. 植物学报 45, 579-587. |
| [5] | 吴文凯, 刘成前, 周志刚, 卢山 (2009). 用于莱茵衣藻荧光定量PCR分析的内参基因选择. 植物生理学通讯 45, 667-672. |
| [6] |
Andersen CL, Jensen JL, ørntoft TF (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets.Cancer Res 64, 5245-5250.
DOI URL PMID |
| [7] |
Bustin SA (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays.J Mol Endocrinol 25, 169-193.
DOI URL |
| [8] |
Bustin SA (2010). Why the need for qPCR publication guidelines?—The case for MIQE.Methods 50, 217-226.
DOI URL PMID |
| [9] |
Chandel M, Kumar M, Sharma U, Kumar N, Singh B, Kaur S (2016). Isolation and characterization of flavanols fromAnthocephalus cadamba and evaluation of their antioxidant, antigenotoxic, cytotoxic and COX-2 inhibitory activities. Rev Bras Farmacogn 26, 474-483.
DOI URL |
| [10] |
Chen L, Zhong HY, Kuang JF, Li JG, Lu WJ, Chen JY (2011). Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions.Planta 234, 377-390.
DOI URL PMID |
| [11] |
Die JV, Roman B, Flores F, Rowland LJ (2016). Design and sampling plan optimization for RT-qPCR experiments in plants: a case study in blueberry.Front Plant Sci 7, 271.
DOI URL PMID |
| [12] |
Galeano E, Vasconcelos TS, Ramiro DA, de Fátima De Martin V, Carrer H (2014). Identification and validation of quantitative real-time reverse transcription PCR reference genes for gene expression analysis in teak (Tectona grandis L.f.). BMC Res Notes 7, 464.
DOI URL PMID |
| [13] |
Gibson UE, Heid CA, Williams PM (1996). A novel method for real time quantitative RT-PCR.Genome Res 6, 995-1001.
DOI URL |
| [14] |
Ginzinger DG (2002). Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream.Exp Hematol 30, 503-512.
DOI URL |
| [15] |
Ho WS, Pang SL, Abdullah J (2014). Identification and analysis of expressed sequence tags present in xylem tissues of kelampayan (Neolamarckia cadamba(Roxb.) Bosser). Physiol Mol Biol Plants 20, 393-397.
DOI URL PMID |
| [16] |
Jian B, Liu B, Bi YR, Hou WS, Wu CX, Han TF (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR.BMC Mol Biol 9, 59.
DOI URL PMID |
| [17] | Li JC, Hu XS, Huang XL, Huo HQ, Li JJ, Zhang D, Li P, Ouyang KX, Chen XY (2017). Functional identification of an EXPA gene(NcEXPA8) isolated from the tree Neolamarckia cadamba. Biotechnol Biotec Eq 31, 1116-1125. |
| [18] |
Liu ZL, Slininger PJ (2007). Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR.J Microbiol Meth 68, 486-496.
DOI URL PMID |
| [19] |
Martins MQ, Fortunato AS, Rodrigues WP, Partelli FL, Campostrini E, Lidon FC, DaMatta FM, Ramalho JC, Ribeiro-Barros AI (2017). Selection and validation of reference genes for accurate RT-qPCR data normalization in Coffea spp. under a climate changes context of interacting elevated [CO2] and temperature. Front Plant Sci 8, 307.
DOI URL PMID |
| [20] | Ouyang KX, Li JC, Huang H, Que QM, Li P, Chen XY (2014). A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. Biotechnol Biotec Eq 28, 1008-1013. |
| [21] |
Ouyang KX, Li JC, Zhao XH, Que QM, Li P, Huang H, Deng XM, Singh SK, Wu AM, Chen XY (2016). Transcriptomic analysis of multipurpose timber yielding tree Neolamarckia cadamba during xylogenesis using RNA- Seq. PLoS One 11, e0159407.
DOI URL PMID |
| [22] |
Pandey A, Negi PS (2016). Traditional uses, phytochemistry and pharmacological properties of Neolamarckia cad- amba: a review. J Ethnopharmacol 181, 118-135.
DOI URL PMID |
| [23] | Pandey A, Negi PS (2018). Phytochemical composition,in vitro antioxidant activity and antibacterial mechanisms of Neolamarckia cadamba fruits extracts. Nat Prod Res 32, 1189-1192. |
| [24] |
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper- Excel-based tool using pair-wise correlations.Biotechnol Lett 26, 509-515.
DOI URL |
| [25] |
Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999). Housekeeping genes as internal standards: use and limits.J Biotechnol 75, 291-295.
DOI URL |
| [26] | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, research0034. |
| [27] |
Zhao XH, Ouyang KX, Gan SM, Zeng W, Song LL, Zhao S, Li JC, Doblin MS, Bacic A, Chen XY, Marchant A, Deng XM, Wu AM (2014). Biochemical and molecular changes associated with heteroxylan biosynthesis in Neolamarckia cadamba(Rubiaceae) during xylogenesis. Front Plant Sci 5, 602.
DOI URL PMID |
| [28] |
Zhou X, Tarver MR, Bennett GW, Oi FM, Scharf ME (2006). Two hexamerin genes from the termite Reticulitermes flavipes: sequence, expression, and proposed functions in caste regulation. Gene 376, 47-58.
DOI URL PMID |
| [1] | Qiong Jiang, Youning Wang, Lixiang Wang, Zhengxi Sun, Xia Li. Validation of Reference Genes for Quantitative RT-PCR Analysis in Soybean Root Tissue under Salt Stress [J]. Chinese Bulletin of Botany, 2015, 50(6): 754-764. |
| [2] | Xiaojuan Su, Baoguo Fan, Lichai Yuan, Xiuna Cui, Shanfa Lu. Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis of Gene Expression in Populus trichocarpa [J]. Chinese Bulletin of Botany, 2013, 48(5): 507-518. |
| [3] | Ran Li, Jiancai Li, Guoxin Zhou, Yonggen Lou. Validation of Rice Candidate Reference Genes for Herbivore-induced Quantitative Real-time PCR Analysis [J]. Chinese Bulletin of Botany, 2013, 48(2): 184-191. |
| [4] | Wei Yuan, Hongjian Wan, Yuejian Yang. Characterization and Selection of Reference Genes for Real-time Quantitative RT-PCR of Plants [J]. Chinese Bulletin of Botany, 2012, 47(4): 427-436. |
| [5] | Meilian Sun, Yunsheng Wang, Dongqing Yang, Chaoling Wei, Liping Gao, Tao Xia, Yu Shan, Yang Luo. Reference Genes for Real-time Fluorescence Quantitative PCR in Camellia sinensis [J]. Chinese Bulletin of Botany, 2010, 45(05): 579-587. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||