

Chinese Bulletin of Botany ›› 2025, Vol. 60 ›› Issue (2): 218-234.DOI: 10.11983/CBB24098 cstr: 32102.14.CBB24098
• EXPERIMENTAL COMMUNICATION • Previous Articles Next Articles
Zhigang Yang1,*,†( ), Pengcheng Zhang1,†, Haiwen Chang1, Liru Kang1, Yi Zuo2, Haoxin Xiang2, Fengying Han1
), Pengcheng Zhang1,†, Haiwen Chang1, Liru Kang1, Yi Zuo2, Haoxin Xiang2, Fengying Han1
Received:2024-06-28
Accepted:2025-01-20
Online:2025-03-10
Published:2025-01-21
Contact:
Zhigang Yang
About author:First author contact:†These authors contributed equally to this paper
Zhigang Yang, Pengcheng Zhang, Haiwen Chang, Liru Kang, Yi Zuo, Haoxin Xiang, Fengying Han. Genetic Diversity Analysis of Pepper Germplasms Based on Morphological Traits and SSR Markers[J]. Chinese Bulletin of Botany, 2025, 60(2): 218-234.
| Primer number | Primer name | Fluorescence labeling | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|---|
| SPE13293 | SPE13293_Es330 | 5′-ROX | GTAGCCATGGCAGAATTGGAAG | TTCAGCAGGTTCTGGTTCTGGT |
| SPE13295 | SPE13295_Esl05 | 5′-HEX | CGCATCTACATCAAGAATCAACCA | GATGTAGAACAAGGAAGCAGGGG |
| SPE13297 | SPE13297_Es363 | 5′-TAMRA | GAGGAATTTTGGAGCCACACAC | AGGTGAAATGGGCAGTGGTAGA |
| SPE13299 | SPE13299_Epms-923 | 5′-FAM | CAAAACCAAATAGGTCCCCC | CGCGCAATAATTCAATATCG |
| SPE13301 | SPE13301_Hpmsl-214 | 5′-TAMRA | TGCGAGTACCGAGTTCTTTCTAG | GGCAGTCCTGGGACAACTCG |
| SPE13303 | SPE13303_Epms697 | 5′-ROX | ATGTCGCTCGCAATTTCACT | CGTAGGGAGGAGCGATAGAG |
| SPE13305 | SPE13305_Es201 | 5′-TAMRA | CGGCCAAATTATTTTTCCCAGT | TTCCCATAGTTGAAGAGCCTGC |
| SPE13307 | SPE13307_Es110 | 5′-HEX | TACGGTTCCTCATCCCAGAAGA | AGGATGACCAATGTTTTGCCTC |
| SPE13309 | SPE13309_Es212 | 5′-TAMRA | CCCTTCCCTTTTCTCCCTTCTT | CCAGAAACAGCCATTGATGATG |
| SPE13311 | SPE13311_Es120 | 5′-ROX | GCGGCCTTTTGATTCATACAAT | CGTTTTACTGCCCTATCTGCTTG |
| SPE13313 | SPE13313_Es285 | 5′-HEX | GCACGAGGCAACAAAAGAGAAT | CGCTGAGCGAGACAGAGAGAG |
| SPE13315 | SPE13315_Es292 | 5′-TAMRA | TCCCATTTCGCAAGAAAGAAAA | TCCGATACAGCTGCAGTAGAAAAA |
| SPE13317 | SPE13317_Es297 | 5′-FAM | TCAGCGATTAAGAATGCGATTG | CCAAATTGCCCTCTCTCTTCCT |
| SPE13319 | SPE13319_Es395 | 5′-ROX | TGTAATTAATTGAGGTGCGCGA | TCTCTGGTTGACAATTAGGCCC |
| SPE13321 | SPE13321_Hpmsl-106 | 5′-FAM | TCCAAACTACAAGCCTGCCTAACC | TTTTGCATTATTGAGTCCCACAGC |
| SPE13323 | SPE13323_Epms712 | 5′-FAM | CCACAAAGGGTTTAAGCAGC | AAGGCAGGAGCAGAGTTCAA |
| SPE13325 | SPE13325_Es417 | 5′-TAMRA | AATGCGGAGGAAGAAGAGGAAG | TAGTCCTTATCACCGACACGGC |
| SPE13327 | SPE13327_Es321 | 5′-HEX | ACGAGGTCCACTTCCCCATTAT | TTAGAGAAGGAATAACCGGCAGC |
| SPE13329 | SPE13329_Esl67 | 5′-HEX | CGCGATTCGATTGCTAAATCTC | CTAATTTCCCAGTTGCGTCTGC |
| SPE13331 | SPE13331_Es64 | 5′-HEX | TTCGGCTATTTGAACAGAAGCA | TGATCCCTTCTTGTTTGATTTTTG |
| SPE13333 | SPE13333_Esl33 | 5′-ROX | TTTTGGGGTTCAATAAAGCTGTG | TTCAACAAGATCATCAATTCACCA |
| SPE13335 | SPE13335_Hpmsl-5 | 5′-FAM | CCAAACGAACCGATGAACACTC | GACAATGTTGAAAAAGGTGGAAGAC |
Table 1 SSR primer information
| Primer number | Primer name | Fluorescence labeling | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|---|
| SPE13293 | SPE13293_Es330 | 5′-ROX | GTAGCCATGGCAGAATTGGAAG | TTCAGCAGGTTCTGGTTCTGGT |
| SPE13295 | SPE13295_Esl05 | 5′-HEX | CGCATCTACATCAAGAATCAACCA | GATGTAGAACAAGGAAGCAGGGG |
| SPE13297 | SPE13297_Es363 | 5′-TAMRA | GAGGAATTTTGGAGCCACACAC | AGGTGAAATGGGCAGTGGTAGA |
| SPE13299 | SPE13299_Epms-923 | 5′-FAM | CAAAACCAAATAGGTCCCCC | CGCGCAATAATTCAATATCG |
| SPE13301 | SPE13301_Hpmsl-214 | 5′-TAMRA | TGCGAGTACCGAGTTCTTTCTAG | GGCAGTCCTGGGACAACTCG |
| SPE13303 | SPE13303_Epms697 | 5′-ROX | ATGTCGCTCGCAATTTCACT | CGTAGGGAGGAGCGATAGAG |
| SPE13305 | SPE13305_Es201 | 5′-TAMRA | CGGCCAAATTATTTTTCCCAGT | TTCCCATAGTTGAAGAGCCTGC |
| SPE13307 | SPE13307_Es110 | 5′-HEX | TACGGTTCCTCATCCCAGAAGA | AGGATGACCAATGTTTTGCCTC |
| SPE13309 | SPE13309_Es212 | 5′-TAMRA | CCCTTCCCTTTTCTCCCTTCTT | CCAGAAACAGCCATTGATGATG |
| SPE13311 | SPE13311_Es120 | 5′-ROX | GCGGCCTTTTGATTCATACAAT | CGTTTTACTGCCCTATCTGCTTG |
| SPE13313 | SPE13313_Es285 | 5′-HEX | GCACGAGGCAACAAAAGAGAAT | CGCTGAGCGAGACAGAGAGAG |
| SPE13315 | SPE13315_Es292 | 5′-TAMRA | TCCCATTTCGCAAGAAAGAAAA | TCCGATACAGCTGCAGTAGAAAAA |
| SPE13317 | SPE13317_Es297 | 5′-FAM | TCAGCGATTAAGAATGCGATTG | CCAAATTGCCCTCTCTCTTCCT |
| SPE13319 | SPE13319_Es395 | 5′-ROX | TGTAATTAATTGAGGTGCGCGA | TCTCTGGTTGACAATTAGGCCC |
| SPE13321 | SPE13321_Hpmsl-106 | 5′-FAM | TCCAAACTACAAGCCTGCCTAACC | TTTTGCATTATTGAGTCCCACAGC |
| SPE13323 | SPE13323_Epms712 | 5′-FAM | CCACAAAGGGTTTAAGCAGC | AAGGCAGGAGCAGAGTTCAA |
| SPE13325 | SPE13325_Es417 | 5′-TAMRA | AATGCGGAGGAAGAAGAGGAAG | TAGTCCTTATCACCGACACGGC |
| SPE13327 | SPE13327_Es321 | 5′-HEX | ACGAGGTCCACTTCCCCATTAT | TTAGAGAAGGAATAACCGGCAGC |
| SPE13329 | SPE13329_Esl67 | 5′-HEX | CGCGATTCGATTGCTAAATCTC | CTAATTTCCCAGTTGCGTCTGC |
| SPE13331 | SPE13331_Es64 | 5′-HEX | TTCGGCTATTTGAACAGAAGCA | TGATCCCTTCTTGTTTGATTTTTG |
| SPE13333 | SPE13333_Esl33 | 5′-ROX | TTTTGGGGTTCAATAAAGCTGTG | TTCAACAAGATCATCAATTCACCA |
| SPE13335 | SPE13335_Hpmsl-5 | 5′-FAM | CCAAACGAACCGATGAACACTC | GACAATGTTGAAAAAGGTGGAAGAC |
| Order | Traits | Distribution frequency (%) | Coefficient of variation (%) | Shannon- Wiener diversity index | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||||
| 1 | PS | 4.8 | 14.4 | 80.8 | 19.17 | 0.60 | |||||||||
| 2 | RA | 51.4 | 41.8 | 6.9 | 30.26 | 0.89 | |||||||||
| 3 | MSC | 6.9 | 21.2 | 45.9 | 6.9 | 19.2 | 37.03 | 1.37 | |||||||
| 4 | SV | 60.3 | 26.0 | 11.0 | 2.7 | 141.78 | 1.00 | ||||||||
| 5 | MA | 21.9 | 39.7 | 38.4 | 24.06 | 1.07 | |||||||||
| 6 | PH | 24.7 | 56.9 | 18.5 | 33.86 | 0.98 | |||||||||
| 7 | CL | 1.4 | 9.6 | 47.3 | 41.8 | 21.14 | 1.00 | ||||||||
| 8 | LM | 93.8 | 6.2 | 22.73 | 0.23 | ||||||||||
| 9 | LSV | 85.6 | 13.0 | 0.7 | 0.7 | 267.58 | 0.47 | ||||||||
| 10 | FF | 89.7 | 10.3 | 27.63 | 0.33 | ||||||||||
| 11 | CC | 99.3 | 0.7 | 8.22 | 0.04 | ||||||||||
| 12 | AC | 0.7 | 2.7 | 5.5 | 15.1 | 40.4 | 35.6 | 20.93 | 1.31 | ||||||
| 13 | SC | 83.6 | 16.4 | 55.98 | 0.45 | ||||||||||
| 14 | NF | 96.6 | 3.4 | 17.64 | 0.15 | ||||||||||
| 15 | SL | 1.4 | 16.4 | 82.2 | 15.26 | 0.52 | |||||||||
| 16 | PP | 30.1 | 48.0 | 21.9 | 37.50 | 1.05 | |||||||||
| 17 | FT | 6.9 | 2.7 | 0.7 | 21.9 | 4.8 | 19.2 | 24.0 | 19.9 | 23.52 | 1.91 | ||||
| 18 | FTS | 82.2 | 12.3 | 3.4 | 2.1 | 49.41 | 0.61 | ||||||||
| 19 | GFC | 6.2 | 78.8 | 15.1 | 8.91 | 0.64 | |||||||||
| 20 | MF | 0.7 | 72.6 | 26.7 | 13.98 | 0.62 | |||||||||
| 21 | FTC | 42.5 | 56.9 | 0.7 | 32.15 | 0.72 | |||||||||
| 22 | FS | 89.0 | 6.9 | 4.1 | 39.98 | 0.42 | |||||||||
| 23 | FSR | 69.9 | 28.1 | 2.1 | 158.75 | 0.69 | |||||||||
| 24 | FBC | 7.5 | 43.2 | 49.3 | 26.05 | 0.91 | |||||||||
| 25 | FSC | 45.2 | 47.3 | 5.5 | 2.1 | 105.96 | 0.95 | ||||||||
Table 2 Variation pattern of qualitative traits of 146 pepper germplasms
| Order | Traits | Distribution frequency (%) | Coefficient of variation (%) | Shannon- Wiener diversity index | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||||
| 1 | PS | 4.8 | 14.4 | 80.8 | 19.17 | 0.60 | |||||||||
| 2 | RA | 51.4 | 41.8 | 6.9 | 30.26 | 0.89 | |||||||||
| 3 | MSC | 6.9 | 21.2 | 45.9 | 6.9 | 19.2 | 37.03 | 1.37 | |||||||
| 4 | SV | 60.3 | 26.0 | 11.0 | 2.7 | 141.78 | 1.00 | ||||||||
| 5 | MA | 21.9 | 39.7 | 38.4 | 24.06 | 1.07 | |||||||||
| 6 | PH | 24.7 | 56.9 | 18.5 | 33.86 | 0.98 | |||||||||
| 7 | CL | 1.4 | 9.6 | 47.3 | 41.8 | 21.14 | 1.00 | ||||||||
| 8 | LM | 93.8 | 6.2 | 22.73 | 0.23 | ||||||||||
| 9 | LSV | 85.6 | 13.0 | 0.7 | 0.7 | 267.58 | 0.47 | ||||||||
| 10 | FF | 89.7 | 10.3 | 27.63 | 0.33 | ||||||||||
| 11 | CC | 99.3 | 0.7 | 8.22 | 0.04 | ||||||||||
| 12 | AC | 0.7 | 2.7 | 5.5 | 15.1 | 40.4 | 35.6 | 20.93 | 1.31 | ||||||
| 13 | SC | 83.6 | 16.4 | 55.98 | 0.45 | ||||||||||
| 14 | NF | 96.6 | 3.4 | 17.64 | 0.15 | ||||||||||
| 15 | SL | 1.4 | 16.4 | 82.2 | 15.26 | 0.52 | |||||||||
| 16 | PP | 30.1 | 48.0 | 21.9 | 37.50 | 1.05 | |||||||||
| 17 | FT | 6.9 | 2.7 | 0.7 | 21.9 | 4.8 | 19.2 | 24.0 | 19.9 | 23.52 | 1.91 | ||||
| 18 | FTS | 82.2 | 12.3 | 3.4 | 2.1 | 49.41 | 0.61 | ||||||||
| 19 | GFC | 6.2 | 78.8 | 15.1 | 8.91 | 0.64 | |||||||||
| 20 | MF | 0.7 | 72.6 | 26.7 | 13.98 | 0.62 | |||||||||
| 21 | FTC | 42.5 | 56.9 | 0.7 | 32.15 | 0.72 | |||||||||
| 22 | FS | 89.0 | 6.9 | 4.1 | 39.98 | 0.42 | |||||||||
| 23 | FSR | 69.9 | 28.1 | 2.1 | 158.75 | 0.69 | |||||||||
| 24 | FBC | 7.5 | 43.2 | 49.3 | 26.05 | 0.91 | |||||||||
| 25 | FSC | 45.2 | 47.3 | 5.5 | 2.1 | 105.96 | 0.95 | ||||||||
| Order | Traits | Means ± SD | Range | Coefficient of variation (%) | Shannon-Wiener diversity index |
|---|---|---|---|---|---|
| 1 | LB (cm) | 13.00±2.76 | 7.94-22.88 | 21.25 | 2.02 |
| 2 | BW (cm) | 6.40±1.63 | 3.42-11.70 | 25.48 | 2.06 |
| 3 | PL (cm) | 6.87±1.92 | 3.22-12.70 | 27.95 | 1.98 |
| 4 | FFN | 10.45±2.78 | 5.00-18.00 | 26.62 | 2.00 |
| 5 | FRL (cm) | 7.92±1.16 | 5.08-11.87 | 14.70 | 1.91 |
| 6 | FW (cm) | 6.96±1.00 | 4.61-10.39 | 14.35 | 1.94 |
| 7 | FRW (g) | 19.82±14.37 | 1.86-80.00 | 72.51 | 1.84 |
| 8 | FTH (cm) | 0.25±0.08 | 0.09-0.23 | 34.04 | 1.93 |
| 9 | LN | 2.28±0.36 | 2.00-3.60 | 15.97 | 1.58 |
Table 3 Variation pattern of quantitative traits of 146 pepper germplasms
| Order | Traits | Means ± SD | Range | Coefficient of variation (%) | Shannon-Wiener diversity index |
|---|---|---|---|---|---|
| 1 | LB (cm) | 13.00±2.76 | 7.94-22.88 | 21.25 | 2.02 |
| 2 | BW (cm) | 6.40±1.63 | 3.42-11.70 | 25.48 | 2.06 |
| 3 | PL (cm) | 6.87±1.92 | 3.22-12.70 | 27.95 | 1.98 |
| 4 | FFN | 10.45±2.78 | 5.00-18.00 | 26.62 | 2.00 |
| 5 | FRL (cm) | 7.92±1.16 | 5.08-11.87 | 14.70 | 1.91 |
| 6 | FW (cm) | 6.96±1.00 | 4.61-10.39 | 14.35 | 1.94 |
| 7 | FRW (g) | 19.82±14.37 | 1.86-80.00 | 72.51 | 1.84 |
| 8 | FTH (cm) | 0.25±0.08 | 0.09-0.23 | 34.04 | 1.93 |
| 9 | LN | 2.28±0.36 | 2.00-3.60 | 15.97 | 1.58 |
| Traits | LB | BW | PL | FFN | FRL | FW | FRW | FTH | LN |
|---|---|---|---|---|---|---|---|---|---|
| LB | 1.00 | ||||||||
| BW | 0.88** | 1.00 | |||||||
| PL | 0.72** | 0.72** | 1.00 | ||||||
| FFN | 0.23** | 0.10 | 0.28** | 1.00 | |||||
| FRL | -0.28** | -0.29** | -0.23** | -0.07 | 1.00 | ||||
| FW | 0.23** | 0.31** | 0.04 | -0.26** | -0.10 | 1.00 | |||
| FRW | 0.18* | 0.22** | 0.05 | -0.25** | 0.24** | 0.81** | 1.00 | ||
| FTH | 0.14 | 0.21* | 0.04 | -0.32** | 0.05 | 0.71** | 0.80** | 1.00 | |
| LN | 0.13 | 0.15 | 0.06 | 0.02 | 0.06 | 0.47** | 0.42** | 0.31** | 1.00 |
Table 4 Correlation coefficient of quantitative traits of 146 pepper germplasms
| Traits | LB | BW | PL | FFN | FRL | FW | FRW | FTH | LN |
|---|---|---|---|---|---|---|---|---|---|
| LB | 1.00 | ||||||||
| BW | 0.88** | 1.00 | |||||||
| PL | 0.72** | 0.72** | 1.00 | ||||||
| FFN | 0.23** | 0.10 | 0.28** | 1.00 | |||||
| FRL | -0.28** | -0.29** | -0.23** | -0.07 | 1.00 | ||||
| FW | 0.23** | 0.31** | 0.04 | -0.26** | -0.10 | 1.00 | |||
| FRW | 0.18* | 0.22** | 0.05 | -0.25** | 0.24** | 0.81** | 1.00 | ||
| FTH | 0.14 | 0.21* | 0.04 | -0.32** | 0.05 | 0.71** | 0.80** | 1.00 | |
| LN | 0.13 | 0.15 | 0.06 | 0.02 | 0.06 | 0.47** | 0.42** | 0.31** | 1.00 |
| Traits | Principal component | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 | PC10 | PC11 | |
| FW | 0.876 | -0.315 | -0.119 | 0.048 | -0.052 | 0.039 | 0.106 | 0.105 | 0.008 | 0.053 | 0.055 |
| FBC | -0.744 | 0.184 | 0.023 | -0.455 | 0.041 | 0.037 | -0.149 | -0.038 | 0.095 | 0.009 | -0.031 |
| FSC | 0.712 | -0.234 | 0.005 | 0.487 | -0.033 | -0.161 | 0.145 | 0.095 | -0.128 | 0.001 | -0.019 |
| FRW | 0.688 | -0.430 | 0.008 | -0.287 | -0.122 | 0.050 | 0.225 | 0.268 | 0.090 | 0.092 | 0.057 |
| FTH | 0.658 | -0.262 | -0.151 | -0.266 | -0.306 | 0.072 | 0.190 | 0.237 | 0.188 | 0.099 | -0.007 |
| FT | -0.626 | -0.042 | 0.133 | -0.397 | -0.034 | -0.031 | 0.018 | 0.034 | 0.059 | 0.056 | 0.045 |
| LN | 0.512 | -0.295 | 0.320 | -0.039 | 0.060 | -0.021 | 0.043 | -0.291 | 0.237 | -0.019 | 0.277 |
| FTC | 0.506 | 0.319 | -0.391 | 0.167 | 0.042 | -0.243 | 0.039 | -0.129 | 0.143 | 0.160 | -0.043 |
| SL | -0.502 | 0.065 | -0.115 | 0.176 | -0.024 | -0.173 | 0.287 | 0.095 | 0.111 | 0.463 | 0.043 |
| MA | -0.065 | 0.714 | 0.220 | 0.007 | -0.256 | -0.009 | 0.192 | 0.141 | -0.026 | -0.143 | -0.023 |
| LB | 0.478 | 0.628 | 0.354 | -0.152 | 0.092 | -0.010 | 0.182 | 0.158 | -0.096 | 0.062 | -0.021 |
| PL | 0.344 | 0.623 | 0.357 | -0.179 | 0.128 | 0.258 | 0.015 | 0.085 | -0.166 | 0.056 | -0.028 |
| BW | 0.585 | 0.606 | 0.192 | -0.256 | 0.126 | 0.030 | 0.125 | 0.021 | -0.100 | 0.081 | -0.012 |
| PS | 0.212 | 0.528 | 0.056 | -0.100 | -0.114 | -0.027 | 0.066 | 0.064 | 0.476 | -0.333 | 0.308 |
| FRL | -0.298 | -0.525 | 0.113 | -0.455 | 0.222 | 0.074 | 0.230 | 0.189 | -0.028 | -0.010 | 0.119 |
| PH | -0.343 | -0.518 | 0.260 | 0.141 | -0.141 | -0.036 | 0.043 | 0.171 | 0.203 | 0.001 | 0.031 |
| FSR | 0.269 | -0.449 | 0.212 | 0.313 | 0.319 | 0.173 | -0.100 | -0.183 | -0.203 | 0.022 | 0.120 |
| CL | -0.339 | 0.404 | -0.194 | 0.330 | 0.101 | 0.239 | 0.063 | 0.234 | 0.029 | -0.003 | 0.157 |
| AC | -0.326 | 0.362 | -0.145 | 0.308 | 0.216 | 0.046 | 0.296 | 0.026 | 0.075 | -0.150 | 0.318 |
| FFN | -0.141 | 0.267 | 0.655 | 0.278 | 0.008 | -0.047 | 0.155 | -0.146 | -0.078 | -0.143 | -0.043 |
| MF | 0.115 | 0.411 | -0.555 | 0.025 | 0.322 | -0.039 | 0.190 | -0.157 | -0.018 | -0.104 | 0.035 |
| FF | 0.318 | 0.117 | 0.554 | -0.115 | -0.025 | -0.083 | -0.080 | -0.262 | 0.204 | 0.191 | -0.238 |
| SV | -0.182 | -0.341 | 0.417 | 0.303 | -0.075 | 0.089 | 0.337 | -0.190 | 0.067 | -0.112 | 0.182 |
| FTS | 0.216 | -0.162 | -0.153 | -0.030 | 0.672 | 0.001 | -0.169 | 0.078 | 0.065 | -0.167 | 0.046 |
| NF | 0.009 | 0.070 | 0.207 | 0.169 | 0.545 | 0.222 | -0.135 | 0.294 | 0.116 | 0.172 | -0.298 |
| GFC | -0.106 | 0.115 | -0.033 | 0.272 | -0.517 | 0.366 | -0.002 | 0.184 | -0.084 | -0.070 | -0.123 |
| RA | 0.186 | -0.107 | -0.100 | -0.051 | 0.201 | 0.694 | 0.001 | 0.157 | 0.112 | -0.086 | -0.014 |
| LSV | -0.328 | -0.271 | 0.167 | 0.002 | -0.010 | 0.477 | 0.310 | -0.104 | -0.100 | 0.068 | 0.036 |
| FS | -0.188 | -0.119 | 0.410 | 0.130 | 0.330 | -0.432 | 0.129 | 0.358 | -0.094 | 0.070 | -0.002 |
| PP | 0.270 | 0.071 | 0.104 | 0.144 | -0.276 | 0.169 | -0.519 | -0.007 | -0.210 | -0.077 | 0.069 |
| SC | -0.125 | 0.003 | -0.296 | -0.032 | -0.062 | 0.053 | 0.512 | -0.109 | -0.318 | 0.089 | -0.260 |
| MSC | -0.197 | 0.119 | 0.047 | 0.391 | -0.117 | -0.013 | -0.224 | 0.407 | 0.287 | 0.236 | -0.004 |
| LM | 0.008 | -0.235 | 0.042 | -0.125 | -0.061 | -0.317 | -0.019 | 0.418 | -0.336 | -0.492 | 0.001 |
| CC | 0.011 | 0.184 | 0.022 | -0.136 | -0.065 | 0.001 | -0.205 | 0.075 | -0.380 | 0.433 | 0.631 |
| Characteristic value | 5.67 | 4.24 | 2.44 | 1.99 | 1.85 | 1.52 | 1.44 | 1.27 | 1.12 | 1.09 | 1.01 |
| Variance contribution rate (%) | 16.66 | 12.46 | 7.18 | 5.86 | 5.43 | 4.47 | 4.23 | 3.74 | 3.29 | 3.21 | 2.98 |
| Cumulative contribution rate (%) | 16.66 | 29.13 | 36.31 | 42.17 | 47.60 | 52.07 | 56.30 | 60.04 | 63.33 | 66.54 | 69.52 |
Table 5 Principal component analysis of pepper germplasm resources
| Traits | Principal component | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 | PC10 | PC11 | |
| FW | 0.876 | -0.315 | -0.119 | 0.048 | -0.052 | 0.039 | 0.106 | 0.105 | 0.008 | 0.053 | 0.055 |
| FBC | -0.744 | 0.184 | 0.023 | -0.455 | 0.041 | 0.037 | -0.149 | -0.038 | 0.095 | 0.009 | -0.031 |
| FSC | 0.712 | -0.234 | 0.005 | 0.487 | -0.033 | -0.161 | 0.145 | 0.095 | -0.128 | 0.001 | -0.019 |
| FRW | 0.688 | -0.430 | 0.008 | -0.287 | -0.122 | 0.050 | 0.225 | 0.268 | 0.090 | 0.092 | 0.057 |
| FTH | 0.658 | -0.262 | -0.151 | -0.266 | -0.306 | 0.072 | 0.190 | 0.237 | 0.188 | 0.099 | -0.007 |
| FT | -0.626 | -0.042 | 0.133 | -0.397 | -0.034 | -0.031 | 0.018 | 0.034 | 0.059 | 0.056 | 0.045 |
| LN | 0.512 | -0.295 | 0.320 | -0.039 | 0.060 | -0.021 | 0.043 | -0.291 | 0.237 | -0.019 | 0.277 |
| FTC | 0.506 | 0.319 | -0.391 | 0.167 | 0.042 | -0.243 | 0.039 | -0.129 | 0.143 | 0.160 | -0.043 |
| SL | -0.502 | 0.065 | -0.115 | 0.176 | -0.024 | -0.173 | 0.287 | 0.095 | 0.111 | 0.463 | 0.043 |
| MA | -0.065 | 0.714 | 0.220 | 0.007 | -0.256 | -0.009 | 0.192 | 0.141 | -0.026 | -0.143 | -0.023 |
| LB | 0.478 | 0.628 | 0.354 | -0.152 | 0.092 | -0.010 | 0.182 | 0.158 | -0.096 | 0.062 | -0.021 |
| PL | 0.344 | 0.623 | 0.357 | -0.179 | 0.128 | 0.258 | 0.015 | 0.085 | -0.166 | 0.056 | -0.028 |
| BW | 0.585 | 0.606 | 0.192 | -0.256 | 0.126 | 0.030 | 0.125 | 0.021 | -0.100 | 0.081 | -0.012 |
| PS | 0.212 | 0.528 | 0.056 | -0.100 | -0.114 | -0.027 | 0.066 | 0.064 | 0.476 | -0.333 | 0.308 |
| FRL | -0.298 | -0.525 | 0.113 | -0.455 | 0.222 | 0.074 | 0.230 | 0.189 | -0.028 | -0.010 | 0.119 |
| PH | -0.343 | -0.518 | 0.260 | 0.141 | -0.141 | -0.036 | 0.043 | 0.171 | 0.203 | 0.001 | 0.031 |
| FSR | 0.269 | -0.449 | 0.212 | 0.313 | 0.319 | 0.173 | -0.100 | -0.183 | -0.203 | 0.022 | 0.120 |
| CL | -0.339 | 0.404 | -0.194 | 0.330 | 0.101 | 0.239 | 0.063 | 0.234 | 0.029 | -0.003 | 0.157 |
| AC | -0.326 | 0.362 | -0.145 | 0.308 | 0.216 | 0.046 | 0.296 | 0.026 | 0.075 | -0.150 | 0.318 |
| FFN | -0.141 | 0.267 | 0.655 | 0.278 | 0.008 | -0.047 | 0.155 | -0.146 | -0.078 | -0.143 | -0.043 |
| MF | 0.115 | 0.411 | -0.555 | 0.025 | 0.322 | -0.039 | 0.190 | -0.157 | -0.018 | -0.104 | 0.035 |
| FF | 0.318 | 0.117 | 0.554 | -0.115 | -0.025 | -0.083 | -0.080 | -0.262 | 0.204 | 0.191 | -0.238 |
| SV | -0.182 | -0.341 | 0.417 | 0.303 | -0.075 | 0.089 | 0.337 | -0.190 | 0.067 | -0.112 | 0.182 |
| FTS | 0.216 | -0.162 | -0.153 | -0.030 | 0.672 | 0.001 | -0.169 | 0.078 | 0.065 | -0.167 | 0.046 |
| NF | 0.009 | 0.070 | 0.207 | 0.169 | 0.545 | 0.222 | -0.135 | 0.294 | 0.116 | 0.172 | -0.298 |
| GFC | -0.106 | 0.115 | -0.033 | 0.272 | -0.517 | 0.366 | -0.002 | 0.184 | -0.084 | -0.070 | -0.123 |
| RA | 0.186 | -0.107 | -0.100 | -0.051 | 0.201 | 0.694 | 0.001 | 0.157 | 0.112 | -0.086 | -0.014 |
| LSV | -0.328 | -0.271 | 0.167 | 0.002 | -0.010 | 0.477 | 0.310 | -0.104 | -0.100 | 0.068 | 0.036 |
| FS | -0.188 | -0.119 | 0.410 | 0.130 | 0.330 | -0.432 | 0.129 | 0.358 | -0.094 | 0.070 | -0.002 |
| PP | 0.270 | 0.071 | 0.104 | 0.144 | -0.276 | 0.169 | -0.519 | -0.007 | -0.210 | -0.077 | 0.069 |
| SC | -0.125 | 0.003 | -0.296 | -0.032 | -0.062 | 0.053 | 0.512 | -0.109 | -0.318 | 0.089 | -0.260 |
| MSC | -0.197 | 0.119 | 0.047 | 0.391 | -0.117 | -0.013 | -0.224 | 0.407 | 0.287 | 0.236 | -0.004 |
| LM | 0.008 | -0.235 | 0.042 | -0.125 | -0.061 | -0.317 | -0.019 | 0.418 | -0.336 | -0.492 | 0.001 |
| CC | 0.011 | 0.184 | 0.022 | -0.136 | -0.065 | 0.001 | -0.205 | 0.075 | -0.380 | 0.433 | 0.631 |
| Characteristic value | 5.67 | 4.24 | 2.44 | 1.99 | 1.85 | 1.52 | 1.44 | 1.27 | 1.12 | 1.09 | 1.01 |
| Variance contribution rate (%) | 16.66 | 12.46 | 7.18 | 5.86 | 5.43 | 4.47 | 4.23 | 3.74 | 3.29 | 3.21 | 2.98 |
| Cumulative contribution rate (%) | 16.66 | 29.13 | 36.31 | 42.17 | 47.60 | 52.07 | 56.30 | 60.04 | 63.33 | 66.54 | 69.52 |
| SSR locus | Number of alleles per locus (Na) | Number of effective alleles per locus (Ne) | Proportion of Ne (%) | Shannon-Wiener diversity index (I) | Observed heterozygosity (Ho) | Expected heterozygosity (He) | Polymorphism information content |
|---|---|---|---|---|---|---|---|
| Epms697 | 3 | 1.847 | 61.57 | 0.701 | 0.082 | 0.459 | 0.365 |
| Epms712 | 3 | 1.981 | 66.03 | 0.738 | 0.068 | 0.495 | 0.386 |
| Epms923 | 6 | 2.282 | 38.03 | 0.965 | 0.082 | 0.562 | 0.469 |
| Es110 | 4 | 1.528 | 38.20 | 0.589 | 0.103 | 0.346 | 0.297 |
| Es120 | 8 | 3.816 | 47.70 | 1.509 | 0.137 | 0.738 | 0.696 |
| Es201 | 4 | 1.383 | 34.58 | 0.525 | 0.048 | 0.277 | 0.250 |
| Es212 | 4 | 1.191 | 29.78 | 0.354 | 0.007 | 0.160 | 0.153 |
| Es285 | 3 | 1.404 | 46.80 | 0.507 | 0.014 | 0.288 | 0.253 |
| Es292 | 3 | 2.067 | 68.90 | 0.863 | 0.041 | 0.516 | 0.446 |
| Es297 | 4 | 1.974 | 49.35 | 0.786 | 0.082 | 0.493 | 0.397 |
| Es321 | 7 | 1.949 | 27.84 | 0.884 | 0.116 | 0.487 | 0.417 |
| Es330 | 3 | 2.061 | 68.70 | 0.889 | 0.062 | 0.515 | 0.461 |
| Es363 | 6 | 2.443 | 40.72 | 1.010 | 0.139 | 0.591 | 0.505 |
| Es395 | 3 | 1.730 | 57.67 | 0.749 | 0.057 | 0.422 | 0.379 |
| Es417 | 5 | 1.380 | 27.60 | 0.557 | 0.041 | 0.275 | 0.253 |
| Es64 | 5 | 1.600 | 32.00 | 0.723 | 0.062 | 0.375 | 0.338 |
| Esl05 | 3 | 1.442 | 48.07 | 0.528 | 0.055 | 0.306 | 0.267 |
| Esl33 | 3 | 1.408 | 46.93 | 0.546 | 0.062 | 0.290 | 0.265 |
| Esl67 | 4 | 2.245 | 56.13 | 0.959 | 0.048 | 0.555 | 0.490 |
| Hpmsl-106 | 2 | 1.914 | 95.70 | 0.671 | 0.048 | 0.478 | 0.364 |
| Hpmsl-214 | 6 | 1.981 | 33.02 | 1.032 | 0.027 | 0.495 | 0.465 |
| Hpmsl-5 | 13 | 5.311 | 40.85 | 2.056 | 0.097 | 0.812 | 0.795 |
| Mean | 4.636 | 2.043 | 44.07 | 0.825 | 0.067 | 0.452 | 0.396 |
Table 6 Polymorphism of 22 pair SSR markers in 146 pepper germplasms
| SSR locus | Number of alleles per locus (Na) | Number of effective alleles per locus (Ne) | Proportion of Ne (%) | Shannon-Wiener diversity index (I) | Observed heterozygosity (Ho) | Expected heterozygosity (He) | Polymorphism information content |
|---|---|---|---|---|---|---|---|
| Epms697 | 3 | 1.847 | 61.57 | 0.701 | 0.082 | 0.459 | 0.365 |
| Epms712 | 3 | 1.981 | 66.03 | 0.738 | 0.068 | 0.495 | 0.386 |
| Epms923 | 6 | 2.282 | 38.03 | 0.965 | 0.082 | 0.562 | 0.469 |
| Es110 | 4 | 1.528 | 38.20 | 0.589 | 0.103 | 0.346 | 0.297 |
| Es120 | 8 | 3.816 | 47.70 | 1.509 | 0.137 | 0.738 | 0.696 |
| Es201 | 4 | 1.383 | 34.58 | 0.525 | 0.048 | 0.277 | 0.250 |
| Es212 | 4 | 1.191 | 29.78 | 0.354 | 0.007 | 0.160 | 0.153 |
| Es285 | 3 | 1.404 | 46.80 | 0.507 | 0.014 | 0.288 | 0.253 |
| Es292 | 3 | 2.067 | 68.90 | 0.863 | 0.041 | 0.516 | 0.446 |
| Es297 | 4 | 1.974 | 49.35 | 0.786 | 0.082 | 0.493 | 0.397 |
| Es321 | 7 | 1.949 | 27.84 | 0.884 | 0.116 | 0.487 | 0.417 |
| Es330 | 3 | 2.061 | 68.70 | 0.889 | 0.062 | 0.515 | 0.461 |
| Es363 | 6 | 2.443 | 40.72 | 1.010 | 0.139 | 0.591 | 0.505 |
| Es395 | 3 | 1.730 | 57.67 | 0.749 | 0.057 | 0.422 | 0.379 |
| Es417 | 5 | 1.380 | 27.60 | 0.557 | 0.041 | 0.275 | 0.253 |
| Es64 | 5 | 1.600 | 32.00 | 0.723 | 0.062 | 0.375 | 0.338 |
| Esl05 | 3 | 1.442 | 48.07 | 0.528 | 0.055 | 0.306 | 0.267 |
| Esl33 | 3 | 1.408 | 46.93 | 0.546 | 0.062 | 0.290 | 0.265 |
| Esl67 | 4 | 2.245 | 56.13 | 0.959 | 0.048 | 0.555 | 0.490 |
| Hpmsl-106 | 2 | 1.914 | 95.70 | 0.671 | 0.048 | 0.478 | 0.364 |
| Hpmsl-214 | 6 | 1.981 | 33.02 | 1.032 | 0.027 | 0.495 | 0.465 |
| Hpmsl-5 | 13 | 5.311 | 40.85 | 2.056 | 0.097 | 0.812 | 0.795 |
| Mean | 4.636 | 2.043 | 44.07 | 0.825 | 0.067 | 0.452 | 0.396 |
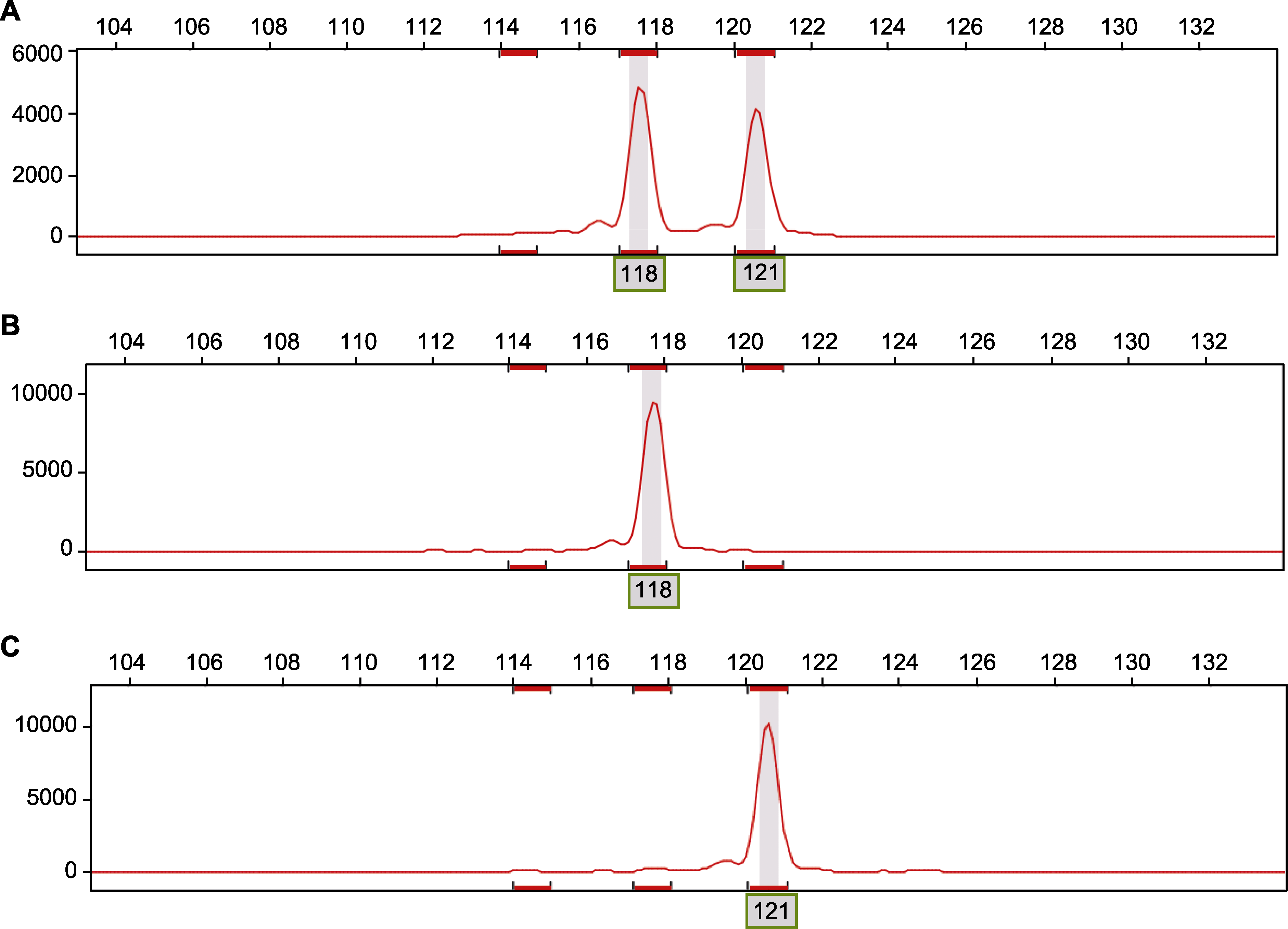
Figure 3 Sequencing of amplification products of samples at the SSR locus Epms697 (A) Heterozygous genotype AB (118 bp/121 bp); (B) Homozygous genotype AA (118 bp); (C) Homozygous genotype BB (121 bp)
| [1] |
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14, 2611-2620.
DOI PMID |
| [2] | Feng PL, Han R, Wang YY, Shao DK, Li QH, Zhong QW (2022). Construction of DNA fingerprints of pepper (Capsicum annuum L.) varieties based on SSR markers. Acta Agric Boreali-Occident Sin 31, 320-327. (in Chinese) |
| 冯鹏龙, 韩睿, 王亚艺, 邵登魁, 李全辉, 钟启文 (2022). 基于SSR标记辣椒品种DNA指纹图谱的构建. 西北农业学报 31, 320-327. | |
| [3] | Fu HF, Lü XH, Chen JY, Li GJ (2018). Genetic diversity analysis of Capsicum germplasm based on phenotypic traits with SSR markers. J Nucl Agric Sci 32, 1309-1319. (in Chinese) |
|
傅鸿妃, 吕晓菡, 陈建瑛, 李国景 (2018). 辣椒种质表型性状与SSR分子标记的遗传多样性分析. 核农学报 32, 1309-1319.
DOI |
|
| [4] | He JL, Shi TT, Chen L, Wang HG, Gao ZJ, Yang MH, Wang RY, Qiao ZJ (2019). The genetic diversity of common millet (Panicum miliaceum) germplasm resources based on the EST-SSR markers. Chin Bull Bot 54, 723-732. (in Chinese) |
|
何杰丽, 石甜甜, 陈凌, 王海岗, 高志军, 杨美红, 王瑞云, 乔治军 (2019). 糜子EST-SSR分子标记的开发及种质资源遗传多样性分析. 植物学报 54, 723-732.
DOI |
|
| [5] | Hu LP (2015). SAS Common Statistical Analysis Tutorial. Beijing: Publishing House of Electronics Industry. pp. 203-452. (in Chinese) |
| 胡良平 (2015). SAS常用统计分析教程. 北京: 电子工业出版社. pp. 203-452. | |
| [6] | Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, Seo E, Choi J, Cheong K, Kim KT, Jung K, Lee GW, Oh SK, Bae C, Kim SB, Lee HY, Kim SY, Kim MS, Kang BC, Jo YD, Yang HB, Jeong HJ, Kang WH, Kwon JK, Shin C, Lim JY, Park JH, Huh JH, Kim JS, Kim BD, Cohen O, Paran I, Suh MC, Lee SB, Kim YK, Shin Y, Noh SJ, Park J, Seo YS, Kwon SY, Kim HA, Park JM, Kim HJ, Choi SB, Bosland PW, Reeves G, Jo SH, Lee BW, Cho HT, Choi HS, Lee MS, Yu Y, Do Choi Y, Park BS, van Deynze A, Ashrafi H, Hill T, Kim WT, Pai HS, Ahn HK, Yeam I, Giovannoni JJ, Rose JKC, Sørensen I, Lee SJ, Kim RW, Choi IY, Choi BS, Lim JS, Lee YH, Choi D (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46, 270-278. |
| [7] |
Lei ML, Liu X, Wang YZ, Cui GQ, Mu ZX, Liu LL, Li X, Lu LH, Li XL, Zhang XJ (2024). Genetic diversity analysis of winter wheat germplasm resources in Shanxi province based on 55K SNP array. Sci Agric Sin 57, 1845-1856. (in Chinese)
DOI |
|
雷梦林, 刘霞, 王艳珍, 崔国庆, 穆志新, 刘龙龙, 李欣, 逯腊虎, 李晓丽, 张晓军 (2024). 基于55K SNP芯片的山西冬小麦种质资源遗传多样性分析. 中国农业科学 57, 1845-1856.
DOI |
|
| [8] | Li XX, Zhang BX (2006). Descriptors and Data Standard for Capsicum. Beijing: Chinese Agriculture Publishing House. pp. 1-98. (in Chinese) |
| 李锡香, 张宝玺 (2006). 辣椒种质资源描述规范和数据标准. 北京: 中国农业出版社. pp. 1-98. | |
| [9] | Liu F, Zhao JT, Sun HH, Xiong C, Sun XP, Wang X, Wang ZY, Jarret R, Wang J, Tang BQ, Xu H, Hu BW, Suo H, Yang BZ, Ou LJ, Li XF, Zhou SD, Yang S, Liu ZB, Yuan F, Pei ZM, Ma YQ, Dai XZ, Wu S, Fei ZJ, Zou XX (2023). Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nat Commun 14, 5487. |
| [10] |
Liu KJ, Muse SV (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128-2129.
DOI PMID |
| [11] | Liu LY, Huang YC, Yang N, Liu WX, Zhao YM, Long CF, Dang XM (2023). Genetic diversity of phenotypic traits in 81 Capsicum annuum germplasms. Chin J Trop Crops 44, 706-715. (in Chinese) |
|
刘林娅, 黄亚成, 杨那, 刘维侠, 赵艳妹, 龙彩凤, 党选民 (2023). 81份辣椒种质资源表型性状的遗传多样性分析. 热带作物学报 44, 706-715.
DOI |
|
| [12] | Liu SR, Yang Y, Tian HL, Yi HM, Wang L, Kang DM, Fan YM, Ren J, Jiang B, Ge JR, Cheng GL, Wang FG (2021). Genetic diversity analysis of silage corn varieties based on agronomic and quality traits and SSR markers. Acta Agron Sin 47, 2362-2370. (in Chinese) |
|
刘少荣, 杨扬, 田红丽, 易红梅, 王璐, 康定明, 范亚明, 任洁, 江彬, 葛建镕, 成广雷, 王凤格 (2021). 基于农艺及品质性状与SSR标记的青贮玉米品种遗传多样性分析. 作物学报 47, 2362-2370.
DOI |
|
| [13] | Lü W, Han JM, Ren GX, Wen F, Wang RP, Liu WP (2021). Genetic diversity analysis of sesame germplasm resources in Shanxi with SSR markers. J Nucl Agric Sci 35, 1495-1506. (in Chinese) |
|
吕伟, 韩俊梅, 任果香, 文飞, 王若鹏, 刘文萍 (2021). 山西芝麻种质资源SSR遗传多样性及群体结构分析. 核农学报 35, 1495-1506.
DOI |
|
| [14] | Pei HX, Wu XX, Feng HP, Gao JX (2024). Genetic diversity analysis of morphological traits of 1000 pepper germplasms. Jiangsu Agric Sci 52(6), 173-190. (in Chinese) |
| 裴红霞, 武旭霞, 冯海萍, 高晶霞 (2024). 1000份辣椒种质形态学性状的遗传多样性分析. 江苏农业科学 52(6), 173-190. | |
| [15] |
Qin C, Yu CS, Shen YO, Fang XD, Chen L, Min JM, Cheng JW, Zhao SC, Xu M, Luo Y, Yang YL, Wu ZM, Mao LK, Wu HY, Ling-Hu CY, Zhou HK, Lin HJ, González- Morales SG, Trejo-Saavedra DL, Tian H, Tang X, Zhao MJ, Huang ZY, Zhou AW, Yao XM, Cui JJ, Li WQ, Chen Z, Feng YQ, Niu YC, Bi SM, Yang XW, Li WP, Cai HM, Luo XR, Montes-Hernández S, Leyva-González MA, Xiong ZQ, He XJ, Bai LJ, Tan S, Tang XQ, Liu D, Liu JW, Zhang SX, Chen MS, Zhang L, Zhang L, Zhang YC, Liao WQ, Zhang Y, Wang M, Lv XD, Wen B, Liu HJ, Luan HM, Zhang YG, Yang S, Wang XD, Xu JH, Li XQ, Li SC, Wang JY, Palloix A, Bosland PW, Li YR, Krogh A, Rivera-Bustamante RF, Herrera-Estrella L, Yin Y, Yu JP, Hu KL, Zhang ZM (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci USA 111, 5135-5140.
DOI PMID |
| [16] | Wang LH, Zhang BX, Zhang ZH, Cao YC, Yu HL, Feng XG (2021). Status in breeding and production of Capsicum spp. in China during ‘the thirteenth five-year plan’ period and future prospect. China Veg (2), 21-29. (in Chinese) |
| 王立浩, 张宝玺, 张正海, 曹亚从, 于海龙, 冯锡刚 (2021). “十三五”我国辣椒育种研究进展、产业现状及展望. 中国蔬菜 (2), 21-29. | |
| [17] |
Wang XY, Zhang FY, Wan X, Wang CQ, Liu Y, Xiao BZ (2023). Diversity of rice landraces revealed by molecular markers and phenotypic traits. J Plant Genet Resour 24, 636-647. (in Chinese)
DOI |
|
王晓映, 张方玉, 万星, 王成琪, 刘燚, 肖本泽 (2023). 基于分子标记和表型性状的水稻地方品种遗传多样性研究. 植物遗传资源学报 24, 636-647.
DOI |
|
| [18] | Xie LF, Li N, Li Y, Yao MH (2019). Genetic diversity and population structure of eggplant (Solanum melongena) germplasm resources based on SRAP method. Chin Bull Bot 54, 58-63. (in Chinese) |
|
谢立峰, 李宁, 李烨, 姚明华 (2019). 茄子种质遗传多样性及群体结构的SRAP分析. 植物学报 54, 58-63.
DOI |
|
| [19] | Yuan JW, Jia L, Tang J, Zhai YQ, Wang S, Yan CS, Fang L, Zhang QA, Yu FF, Gan DF, Jiang HK (2024). Genetic diversity analysis of phenotypic characters in 275 pepper germplasm resources. J Anhui Agric Univ 51, 244-255. (in Chinese) |
| 袁娟伟, 贾利, 唐菁, 翟永琪, 汪胜, 严从生, 方凌, 张其安, 俞飞飞, 甘德芳, 江海坤 (2024). 275份辣椒种质资源表型性状的遗传多样性分析. 安徽农业大学学报 51, 244-255. | |
| [20] | Zhang QQ, Liang S, Wang Y, Jia L, Fang L, Zhang QA, Jiang HK, Sui YH, Dong YX (2020). Genetic diversity analysis of 57 germplasms of Capsicum annuum based on phenotypic traits and SSR markers. J Trop Subtrop Bot 28, 356-366. (in Chinese) |
| 张强强, 梁赛, 王艳, 贾利, 方凌, 张其安, 江海坤, 隋益虎, 董言香 (2020). 基于表型性状和SSR标记的57份辣椒种质遗传多样性分析. 热带亚热带植物学报 28, 356-366. | |
| [21] | Zhang XL, Zhu LL, Li FZ, Tang XM, Xia YL, You Y, Zhong RC (2023). Evaluation and analysis of agronomic and quality traits of 115 peanut germplasm resources. Acta Agric Zhejiang 35, 2033-2044. (in Chinese) |
|
张小利, 朱灵龙, 李付振, 唐秀梅, 夏友霖, 游宇, 钟瑞春 (2023). 115份花生种质资源农艺与品质性状鉴评及分析. 浙江农业学报 35, 2033-2044.
DOI |
|
| [22] |
Zhang YZ, Zhang XJ, Liang D, Guo Q, Fan XQ, Nie ME, Wang HY, Zhao WB, Du WJ, Liu QS (2023). Genetic diversity analysis and comprehensive evaluation of sorghum breeding materials based on phenotypic traits. Sci Agric Sin 56, 2837-2852. (in Chinese)
DOI |
|
张一中, 张晓娟, 梁笃, 郭琦, 范昕琦, 聂萌恩, 王绘艳, 赵文博, 杜维俊, 柳青山 (2023). 基于表型性状的高粱育种材料遗传多样性分析及综合评价. 中国农业科学 56, 2837-2852.
DOI |
|
| [23] | Zhang ZH, Wang YF, Wu HM, Yu HL, Cao YC, Feng XG, Wang LH (2024). Advandes in genetic mapping and candidate gene analysis of important traits in pepper (Capsicum spp.). Acta Hortic Sin 51, 669-696. (in Chinese) |
| 张正海, 王永富, 吴华茂, 于海龙, 曹亚从, 冯锡刚, 王立浩 (2024). 辣椒重要性状遗传定位与候选基因研究进展. 园艺学报 51, 669-696. | |
| [24] | Zhao YH, Qi SN, Li AX, Zhao YX, Ma XR, Liu YX (2024). Genetic diversity analysis of hawthorn germplasm resources based on fruit phenotypic traits. J Shenyang Agric Univ 55, 395-404. (in Chinese) |
| 赵玉辉, 齐索尼, 李昂轩, 赵迎汐, 马欣然, 刘月学 (2024). 基于果实表型性状的山楂种质资源遗传多样性分析. 沈阳农业大学学报 55, 395-404. | |
| [25] | Zou XX, Zhu F (2022). Origin, evolution and cultivation history of the pepper. Acta Hortic Sin 49, 1371-1381. (in Chinese) |
|
邹学校, 朱凡 (2022). 辣椒的起源、进化与栽培历史. 园艺学报 49, 1371-1381.
DOI |
| [1] | 黄 承玲, Han Li Rong, Ling Qing Hong, Xiong Yang Sheng, Ling Tian Xiao, Xia Guowei Xia Guowei, Ren Chen Zheng, Wei Zhou. Study on genetic conservation of Rhododendron liboense based on SNP molecular markers,a plant species with extremely small populations [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | Zhang Ruli, Li Dezhu, Zhang Yuxiao. Population Genetic Structure and Climate Adaptation Analysis of Brachystachyum densiflorum [J]. Chinese Bulletin of Botany, 2025, 60(3): 407-424. |
| [3] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [4] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [5] | HUANG Ling, WANG Zhen, MA Ze, YANG Fa-Lin, LI Lan, SEREKPAYEV Nurlan, NOGAYEV Adilbek, HOU Fu-Jiang. Effects of long-term grazing and nitrogen addition on the growth of Stipa bungeana population in typical steppe of Loess Plateau [J]. Chin J Plant Ecol, 2024, 48(3): 317-330. |
| [6] | LIU Wei-Hui, SONG Xiao-Yan, CAIRENDUOJIE , DING Lu-Ming, WANG Chang-Ting. Effects of degradation degree on the root morphological traits and biomass of dominant plant species in alpine meadows [J]. Chin J Plant Ecol, 2024, 48(12): 1666-1682. |
| [7] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [8] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [9] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [10] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [11] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [12] | Jiang Wang, Yifan Zhao, Yanfu Qu, Caiwen Zhang, Liang Zhang, Chuanwu Chen, Yanping Wang. A dataset of the morphological, life-history, and ecological traits of snakes in China [J]. Biodiv Sci, 2023, 31(7): 23126-. |
| [13] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [14] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [15] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||