

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (6): 935-945.DOI: 10.11983/CBB23050 cstr: 32102.14.CBB23050
• SPECIAL TOPICS • Previous Articles Next Articles
Yi Li1,2,3,†, Xi Zhang1,2,3,†, Yanhui Yuan1,2,3, Pichang Gong4,*( ), Jinxing Lin1,2,3,*(
), Jinxing Lin1,2,3,*( )
)
Received:2023-04-14
Accepted:2023-08-19
Online:2023-11-01
Published:2023-11-27
Contact:
* E-mail: gongpeichang@ibcas.ac.cn;linjx@ibcas.ac.cn
About author:† These authors contributed equally to this paper
Yi Li, Xi Zhang, Yanhui Yuan, Pichang Gong, Jinxing Lin. Aptamers and Their Applications in Plant Science Researches[J]. Chinese Bulletin of Botany, 2023, 58(6): 935-945.
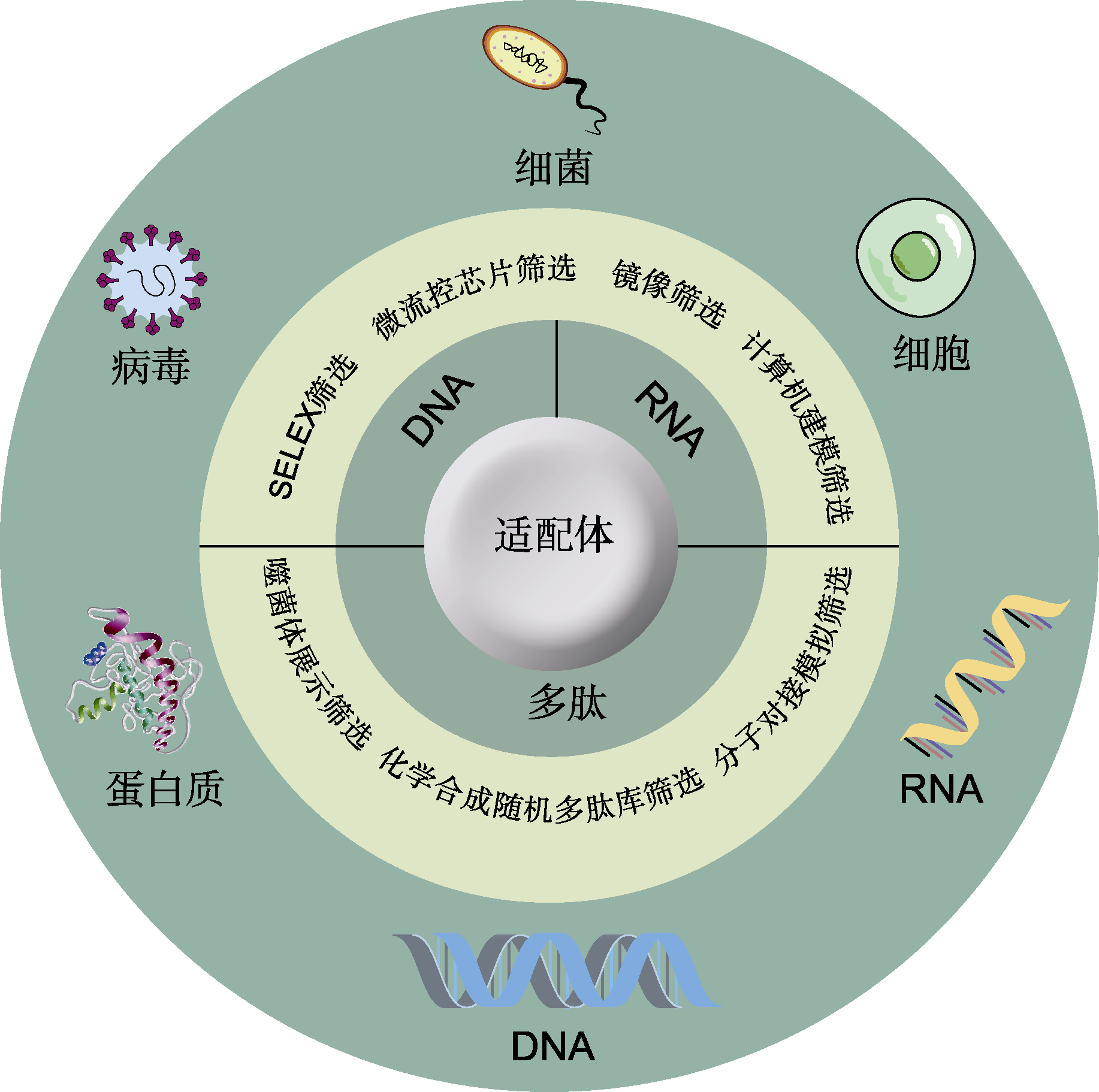
Figure 1 The classification, screening and targets of aptamers The aptamers can be divided into nucleic acid aptamers and peptide aptamers. The nucleic acid aptamers include DNA aptamers and RNA aptamers, and the screening methods mainly include systematic evolution of ligands by exponential enrichment (SELEX) technology, and SELEX based microfluidic chip, mirror-image and computer modeling. Peptide aptamers can be screened by phage display, random peptide library and molecular docking. The targets of aptamers are mainly involved in cells, bacteria, virus, DNA, RNA, and proteins.
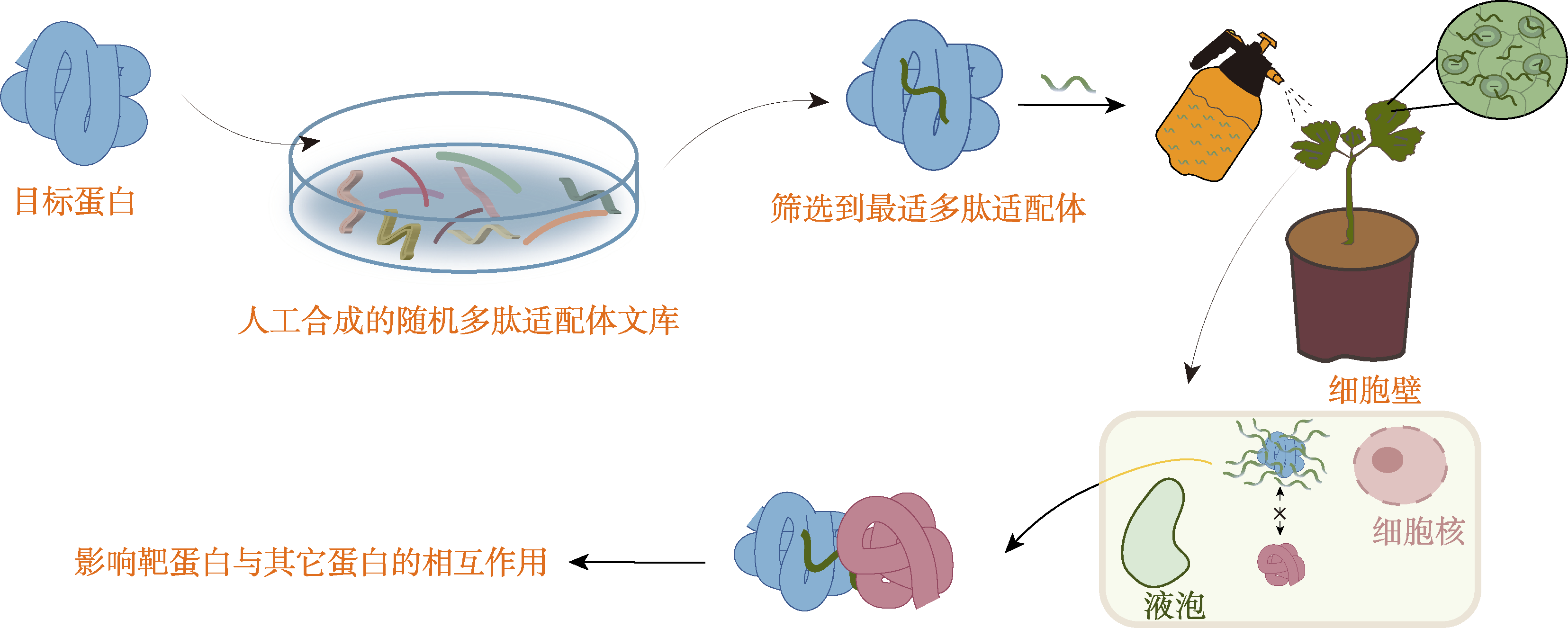
Figure 2 The process of selecting the most suitable peptide aptamer for the target protein through the synthetic peptide aptamer library The optimal peptide aptamer binding to the target protein was screened in vitro by using a synthetic peptide library. Peptide aptamers are sprayed on the surface of plants in vitro, and peptide aptamers enter plant cells to affect the interaction between target proteins and other proteins, thereby inhibiting the function of target proteins.
| [1] | 蔡先洪, 孙强, 吴谦, 叶浩楠, 巫沁芳, 区文卉, 陈锦灿, 陈兰美 (2022). 基于毛细管电泳法-指数富集配体系统进化筛选维吉霉素M1适配体及其检测应用. 分析化学 50, 728-738. |
| [2] | 公丕昌, 王丽, 贺超英 (2010). 多肽配体技术在植物功能基因组学中的应用前景. 遗传 32, 548-554. |
| [3] |
黄子珂, 刘超, 付强强, 李进, 邹建梅, 谢斯滔, 邱丽萍 (2018). 核酸适配体荧光探针在生化分析和生物成像中的研究进展. 应用化学 35, 28-39.
DOI |
| [4] | 林禹晴, 毛东鹏, 胡加枫, 王鑫垚, 段鹏虎, 朴云仙 (2022). 基于甘蔗渣衍生碳量子点的荧光适配体探针的制备及其对17β-雌二醇的检测效果. 环境工程学报 16, 3796-3804. |
| [5] | 梅雪娇, 李梦思, 杨阳, 刘萌, 刘红, 韩欣宇, 曹敏杰, 刘光明 (2019). 拟穴青蟹原肌球蛋白抗原表位适配体小肽的筛选与鉴定. 食品安全质量检测学报 10, 1790-1796. |
| [6] | 覃磊鑫, 刘倩, 闵楠, 边江涛, 高云, 王梁华, 焦炳华, 孙铭娟 (2022). 计算机模拟海洋生物毒素适配体的应用. 生命的化学 42, 1617-1624. |
| [7] |
王建武, 王文娟, 相微微, 代惠萍, 王海庆, 屈香香, 亢福仁 (2022). 过表达MtVP1对马铃薯表型及糖代谢的影响. 植物学报 57, 197-208.
DOI |
| [8] |
王田幸子, 朱峥, 陈悦, 刘玉晴, 燕高伟, 徐珊, 张彤, 马金姣, 窦世娟, 李莉云, 刘国振 (2021). 水稻OsWRKY42是Xa21介导的抗白叶枯病途径新元件. 植物学报 56, 687-698.
DOI |
| [9] |
谢先荣, 曾栋昌, 谭健韬, 祝钦泷, 刘耀光 (2021). 基于CRISPR编辑系统的DNA片段删除技术. 植物学报 56, 44-49.
DOI |
| [10] |
杨凯如, 贾绮玮, 金佳怡, 叶涵斐, 王盛, 陈芊羽, 管易安, 潘晨阳, 辛德东, 方媛, 王跃星, 饶玉春 (2022). 水稻黄绿叶调控基因YGL18的克隆与功能解析. 植物学报 57, 276-287.
DOI |
| [11] |
Aagaard L, Rossi JJ (2007). RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev 59, 75-86.
DOI URL |
| [12] |
Abdeeva IA, Maloshenok LG, Pogorelko GV, Mokrykova MV, Bruskin SA (2019). RNA-aptamers—as targeted inhibitors of protein functions in plants. J Plant Physiol 232, 127-129.
DOI URL |
| [13] |
Apostolopoulos V, Bojarska J, Chai TT, Elnagdy S, Kaczmarek K, Matsoukas J, New R, Parang K, Lopez OP, Parhiz H, Perera CO, Pickholz M, Remko M, Saviano M, Skwarczynski M, Tang YF, Wolf WM, Yoshiya T, Zabrocki J, Zielenkiewicz P, AlKhazindar M, Barriga V, Kelaidonis K, Sarasia EM, Toth I (2021). A global review on short peptides: frontiers and perspectives. Molecules 26, 430.
DOI URL |
| [14] |
Bai JY, Luo Y, Wang X, Li S, Luo M, Yin M, Zuo YL, Li GL, Yao JY, Yang H, Zhang MD, Wei W, Wang ML, Wang R, Fan CH, Zhao Y (2020). A protein-independent fluorescent RNA aptamer reporter system for plant genetic engineering. Nat Commun 11, 3847.
DOI PMID |
| [15] |
Bao ZL, Clancy MA, Carvalho RF, Elliott K, Folta KM (2017). Identification of novel growth regulators in plant populations expressing random peptides. Plant Physiol 175, 619-627.
DOI PMID |
| [16] |
Chen J, Chen MY, Zhu TF (2022). Directed evolution and selection of biostable L-DNA aptamers with a mirror-image DNA polymerase. Nat Biotechnol 40, 1601-1609.
DOI PMID |
| [17] |
Cho JH, Ha NR, Koh SH, Yoon MY (2016). Design of a PKCδ-specific small peptide as a theragnostic agent for glioblastoma. Anal Biochem 496, 63-70.
DOI URL |
| [18] |
Colombo M, Masiero S, Rosa S, Caporali E, Toffolatti SL, Mizzotti C, Tadini L, Rossi F, Pellegrino S, Musetti R, Velasco R, Perazzolli M, Vezzulli S, Pesaresi P (2020). NoPv1: a synthetic antimicrobial peptide aptamer targeting the causal agents of grapevine downy mildew and potato late blight. Sci Rep 10, 17574.
DOI PMID |
| [19] |
Colombo M, Mizzotti C, Masiero S, Kater MM, Pesaresi P (2015). Peptide aptamers: the versatile role of specific protein function inhibitors in plant biotechnology. J Integr Plant Biol 57, 892-901.
DOI |
| [20] |
Ellington AD, Szostak JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818-822.
DOI |
| [21] |
Głazowska S, Mravec J (2021). An aptamer highly specific to cellulose enables the analysis of the association of cellulose with matrix cell wall polymers in vitro and in muro. Plant J 108, 579-599.
DOI URL |
| [22] |
Gong PC, Quan H, He CY (2014). Targeting MAGO proteins with a peptide aptamer reinforces their essential roles in multiple rice developmental pathways. Plant J 80, 905-914.
DOI URL |
| [23] |
Gray BP, Brown KC (2014). Combinatorial peptide libraries: mining for cell-binding peptides. Chem Rev 114, 1020-1081.
DOI PMID |
| [24] |
Hanes J, Schaffitzel C, Knappik A, Plückthun A (2000). Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat Biotechnol 18, 1287-1292.
PMID |
| [25] |
Hao ZZ, Gong PC, He CY, Lin JX (2018). Peptide aptamers to inhibit protein function in plants. Trends Plant Sci 23, 281-284.
DOI PMID |
| [26] |
Hoppe-Seyler F, Crnkovic-Mertens I, Tomai E, Butz K (2004). Peptide aptamers: specific inhibitors of protein function. Curr Mol Med 4, 529-538.
PMID |
| [27] |
Huang JJ, Wang D, Li H, Tang YQ, Ma X, Tang HQ, Lin M, Liu Z (2022). Antifungal activity of an artificial peptide aptamer SNP-D4 against Fusarium oxysporum. PeerJ 10, e12756.
DOI URL |
| [28] |
Ishida R, Adachi T, Yokota A, Yoshihara H, Aoki K, Nakamura Y, Hamada M (2020). RaptRanker: in silico RNA aptamer selection from HT-SELEX experiment based on local sequence and structure information. Nucleic Acids Res 48, e82.
DOI URL |
| [29] |
Islam MZ, Sharmin S, Moniruzzaman M, Yamazaki M (2018). Elementary processes for the entry of cell-penetrating peptides into lipid bilayer vesicles and bacterial cells. Appl Microbiol Biotechnol 102, 3879-3892.
DOI PMID |
| [30] |
Jeong HY, Kim H, Lee M, Hong JJ, Lee JH, Kim J, Choi MJ, Park YS, Kim SC (2020). Development of HER2- specific aptamer-drug conjugate for breast cancer therapy. Int J Mol Sci 21, 9764.
DOI URL |
| [31] |
Kim SH, Lee EH, Lee SC, Kim AR, Park HH, Son JW, Koh SH, Yoon MY (2020). Development of peptide aptamers as alternatives for antibody in the detection of amyloid- beta 42 aggregates. Anal Biochem 609, 113921.
DOI URL |
| [32] |
Lautner G, Balogh Z, Bardóczy V, Mészáros T, Gyurcsányi RE (2010). Aptamer-based biochips for label-free detection of plant virus coat proteins by SPR imaging. Analyst 135, 918-926.
DOI PMID |
| [33] |
Li JY, Zhang C, He YB, Li SY, Yan L, Li YC, Zhu ZW, Xia LQ (2023). Plant base editing and prime editing: the current status and future perspectives. J Integr Plant Biol 65, 444-467.
DOI |
| [34] |
Li Z, Uzawa T, Zhao HC, Luo SC, Yu HH, Kobatake E, Ito Y (2014). In vitro selection of peptide aptamers using a ribosome display for a conducting polymer. J Biosci Bioeng 117, 501-503.
DOI PMID |
| [35] |
Liu CC, Mack AV, Tsao ML, Mills JH, Lee HS, Choe H, Farzan M, Schultz PG, Smider VV (2008). Protein evolution with an expanded genetic code. Proc Natl Acad Sci USA 105, 17688-17693.
DOI PMID |
| [36] |
Liu CX, Chen LL (2022). Circular RNAs: characterization, cellular roles, and applications. Cell 185, 2390.
DOI URL |
| [37] |
Liu DX, Tien TTT, Bao DT, Linh NTP, Park H, Yeo SJ (2019). A novel peptide aptamer to detect Plasmodium falciparum lactate dehydrogenase. J Biomed Nanotechnol 15, 204-211.
DOI URL |
| [38] |
Lou XH, Qian JR, Xiao Y, Viel L, Gerdon AE, Lagally ET, Atzberger P, Tarasow TM, Heeger AJ, Soh HT (2009). Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci USA 106, 2989-2994.
DOI PMID |
| [39] |
Mendoza-Figueroa JS, Kvarnheden A, Méndez-Lozano J, Rodríguez-Negrete EA, de los Monteros RA, Soriano-García M (2018). A peptide derived from enzymatic digestion of globulins from amaranth shows strong affinity binding to the replication origin of tomato yellow leaf curl virus reducing viral replication in Nicotiana benthamiana. Pestic Biochem Physiol 145, 56-65.
DOI PMID |
| [40] |
Millward SW, Fiacco S, Austin RJ, Roberts RW (2007). Design of cyclic peptides that bind protein surfaces with antibody-like affinity. ACS Chem Biol 2, 625-634.
PMID |
| [41] |
Mohr S, Bakal C, Perrimon N (2010). Genomic screening with RNAi: results and challenges. Annu Rev Biochem 79, 37-64.
DOI PMID |
| [42] |
Mou QB, Xue XY, Ma Y, Banik M, Garcia V, Guo WJ, Wang J, Song TJ, Chen LQ, Lu Y (2022). Efficient delivery of a DNA aptamer-based biosensor into plant cells for glucose sensing through thiol-mediated uptake. Sci Adv 8, eabo0902.
DOI URL |
| [43] |
Nelson CE, Gersbach CA (2016). Engineering delivery vehicles for genome editing. Annu Rev Chem Biomol Eng 7, 637-662.
DOI PMID |
| [44] |
Nimjee SM, White RR, Becker RC, Sullenger BA (2017). Aptamers as therapeutics. Annu Rev Pharmacol Toxicol 57, 61-79.
DOI PMID |
| [45] |
Ormancey M, Guillotin B, Merret R, Camborde L, Duboé C, Fabre B, Pouzet C, Impens F, van Haver D, Carpentier MC, Clemente HS, Aguilar M, Lauressergues D, Scharff LB, Pichereaux C, Burlet-Schiltz O, Bousquet- Antonelli C, Gevaert K, Thuleau P, Plaza S, Combier JP (2023). Complementary peptides represent a credible alternative to agrochemicals by activating translation of targeted proteins. Nat Commun 14, 254.
DOI PMID |
| [46] |
Pagadala NS, Syed K, Tuszynski J (2017). Software for molecular docking: a review. Biophys Rev 9, 91-102.
DOI PMID |
| [47] |
Qu J, Yu SQ, Zheng Y, Zheng Y, Yang H, Zhang JL (2017). Aptamer and its applications in neurodegenerative diseases. Cell Mol Life Sci 74, 683-695.
DOI PMID |
| [48] |
Roy S, Narang BK, Gupta MK, Abbot V, Singh V, Rawal RK (2018). Molecular docking studies on isocytosine analogues as xanthine oxidase inhibitors. Drug Res (Stuttg) 68, 395-402.
DOI PMID |
| [49] |
Song CG, Yang J, Wang YD, Ding G, Guo LP, Qin JC (2022). Mechanisms and transformed products of aflatoxin B1 degradation under multiple treatments: a review. Crit Rev Food Sci Nutr 14, 1-13.
DOI URL |
| [50] |
Song XF, Guo P, Ren SC, Xu TT, Liu CM (2013). Antagonistic peptide technology for functional dissection of CLV3/ ESR genes in Arabidopsis. Plant Physiol 161, 1076-1085.
DOI URL |
| [51] |
Tan WH, Donovan MJ, Jiang JH (2013). Aptamers from cell-based selection for bioanalytical applications. Chem Rev 113, 2842-2862.
DOI PMID |
| [52] |
Tan Y, Li YY, Qu YX, Su YY, Peng YB, Zhao ZL, Fu T, Wang XQ, Tan WH (2021). Aptamer-peptide conjugates as targeted chemosensitizers for breast cancer treatment. ACS Appl Mater Interfaces 13, 9436-9444.
DOI URL |
| [53] |
Tan YY, Guo QP, Xie Q, Wang KM, Yuan BY, Zhou Y, Liu JB, Huang J, He XX, Yang XH, He CM, Zhao XY (2014). Single-walled carbon nanotubes (SWCNTs)-assisted cell- systematic evolution of ligands by exponential enrichment (Cell-SELEX) for improving screening efficiency. Anal Chem 86, 9466-9472.
DOI URL |
| [54] |
Torti S, Schlesier R, Thümmler A, Bartels D, Römer P, Koch B, Werner S, Panwar V, Kanyuka K, Wirén NV, Jones JDG, Hause G, Giritch A, Gleba Y (2021). Transient reprogramming of crop plants for agronomic performance. Nat Plants 7, 159-171.
DOI PMID |
| [55] |
Tuerk C, Gold L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505-510.
DOI PMID |
| [56] |
Tungsirisurp S, O’Reilly R, Napier R (2023). Nucleic acid aptamers as aptasensors for plant biology. Trends Plant Sci 28, 359-371.
DOI URL |
| [57] |
Wang HF, La Russa M, Qi LS (2016). CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem 85, 227-264.
DOI PMID |
| [58] |
Wang Q, Liu W, Xing YQ, Yang XH, Wang KM, Jiang R, Wang P, Zhao Q (2014). Screening of DNA aptamers against myoglobin using a positive and negative selection units integrated microfluidic chip and its biosensing application. Anal Chem 86, 6572-6579.
DOI PMID |
| [59] |
Wang T, Chen CY, Larcher LM, Barrero RA, Veedu RN (2019). Three decades of nucleic acid aptamer technologies: lessons learned, progress and opportunities on aptamer development. Biotechnol Adv 37, 28-50.
DOI PMID |
| [60] |
Wilson DS, Keefe AD, Szostak JW (2001). The use of mRNA display to select high-affinity protein-binding peptides. Proc Natl Acad Sci USA 98, 3750-3755.
PMID |
| [61] |
Xiao XR, Li H, Zhao LJ, Zhang YF, Liu ZC (2021). Oligonucleotide aptamers: recent advances in their screening, molecular conformation and therapeutic applications. Biomed Pharmacother 143, 112232.
DOI PMID |
| [62] |
Xu Q, Ye X, Ma X, Li H, Tang HQ, Tang YQ, Liu Z (2019). Engineering a peptide aptamer to target calmodulin for the inhibition of Magnaporthe oryzae. Fungal Biol 123, 489-496.
DOI URL |
| [63] |
Xu ZF, Jiang XL, Li Y, Ma X, Tang YQ, Li H, Yi KX, Li JJ, Liu Z (2022). Antifungal activity of montmorillonite/peptide aptamer nanocomposite against Colletotrichum gloeosporioides on Stylosanthes. Int J Biol Macromol 217, 282-290.
DOI URL |
| [64] |
Yang CH, Tsai CH (2022). Aptamer against Aflatoxin B1 obtained by SELEX and applied in detection. Biosensors 12, 848.
DOI URL |
| [65] |
Zhang J, Lv XF, Feng W, Li XQ, Li KJ, Deng YL (2018). Aptamer-based fluorometric lateral flow assay for creatine kinase MB. Mikrochim Acta 185, 364.
DOI PMID |
| [66] |
Zhang YY, Li LZ, Zhang H, Shang JJ, Li C, Naqvi SMZA, Birech Z, Hu JD (2022a). Ultrasensitive detection of plant hormone abscisic acid-based surface-enhanced Raman spectroscopy aptamer sensor. Anal Bioanal Chem 414, 2757-2766.
DOI |
| [67] |
Zhang ZL, Liu C, Li K, Li XX, Xu MM, Guo YF (2022b). CLE14 functions as a "brake signal" to suppress age-dependent and stress-induced leaf senescence by promoting JUB1-mediated ROS scavenging in Arabidopsis. Mol Plant 15, 179-188.
DOI URL |
| [68] |
Zhou JH, Rossi J (2017). Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov 16, 181-202.
DOI PMID |
| [69] |
Zhou Y, Zhuo YT, Peng RZ, Zhang YT, Du YL, Zhang Q, Sun Y, Qiu LP (2021). Functional nucleic acid-based cell imaging and manipulation. Sci China Chem 64, 1817-1825.
DOI |
| [70] |
Zhu C, Yang G, Ghulam M, Li LS, Qu F (2019). Evolution of multi-functional capillary electrophoresis for high-efficiency selection of aptamers. Biotechnol Adv 37, 107432.
DOI URL |
| [1] | Lu Zi-Jia, Wang Tian-Rui, Zheng Si-Si, Cao Jian-Guo, Kozlowski Gregor, Song Yi-Gang. Environmental adaptive genetic variation and genetic vulnerability of relict plant Pterocarya hupehensis [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | ZHANG Zi-Rui, Zhou Jing, HU Yan-Ping, Liang Shuang, MA Yong-Peng, CHEN Wei-Le. Root-associated Fungal Communities of the Critically Endangered Plant Pinus Squamata [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [3] | Gan Xie, Jing Xuan, Qidi Fu, Ze Wei, Kai Xue, Hairui Luo, Jixi Gao, Min Li. Establishing an intelligent identification model for unmanned aerial vehicle surveys of grassland plant diversity [J]. Biodiv Sci, 2025, 33(4): 24236-. |
| [4] | Tao Xie, Yifan Zhang, Yunhui Liu, Huiyu You, Jibenben Xia, Rong Ma, Chunni Zhang, Xuejun Hua. Research progress of iron-sulfur cluster synthesis system and regulation in plant mitochondria [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [5] | JIANG Xiao-Yu, YU Xin-Miao, LIAO Qin, ZHANG Jin-Wei, WU Xue-Feng, WANG Xu, PAN Jun-Tong, WANG Jun-Feng, MU Chun-Sheng, SHI Yu-Jie. Studies on the emission of nitrous oxide from terrestrial plants [J]. Chin J Plant Ecol, 2025, 49(4): 513-525. |
| [6] | Xiong Lianglin, Liang Guolu, Guo Qigao, Jing Danlong. Advances in the Regulation of Alternative Splicing of Genes in Plants in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2025, 60(3): 435-448. |
| [7] | Zhang Chan, Zhao Suya, Zhang Xinran, Wang Yifan, Wang Linlin. Impacts of alien pollinators on native plant‒pollinator interactions [J]. Biodiv Sci, 2025, 33(2): 24443-. |
| [8] | Yan Deng, Limin Lu, Qiang Zhang, Zhiduan Chen, Haihua Hu. A Comprehensive Evaluation of the Plastid DNA Data Gaps of Vascular Plants in Species and Geographic Area [J]. Chinese Bulletin of Botany, 2025, 60(1): 1-16. |
| [9] | Ruoyue Li, Xiaochao Yang, Zhanqing Hao, Shihong Jia. The intensity of heat waves and insect herbivory on campus plants and their relationship with leaf functional traits [J]. Biodiv Sci, 2025, 33(1): 24283-. |
| [10] | Jing Xuan, Qidi Fu, Gan Xie, Kai Xue, Hairui Luo, Ze Wei, Mingyue Zhao, Liang Zhi, Huawei Wan, Jixi Gao, Min Li. An Artificial Intelligence Model for Identifying Grassland Plants in Northern China [J]. Chinese Bulletin of Botany, 2025, 60(1): 74-80. |
| [11] | Xiangtan Yao, Xinyi Zhang, Yang Chen, Ye Yuan, Wangda Cheng, Tianrui Wang, Yingxiong Qiu. Genomic resequencing reveals the genetic diversity of the cultivated water caltrop, and the origin and domestication of ‘Nanhuling’ [J]. Biodiv Sci, 2024, 32(9): 24212-. |
| [12] | WANG Yi-Tong, Yeerjiang BAIKETUERHAN, LIAO Dan, WANG Juan. Correlation between elemental biometric characteristics and sexual dimorphism in leaves of dioecious Acer barbinerve at different growth stages [J]. Chin J Plant Ecol, 2024, 48(6): 760-769. |
| [13] | Hongju Li, Weicai Yang. A Micropeptide With a Big Role: New Molecular Mechanism in Seed Desiccation [J]. Chinese Bulletin of Botany, 2024, 59(6): 869-872. |
| [14] | Chunjiao Xia, Yunguang Li, Shu Xia, Wei Pang, Chunli Chen. Flow Cytometric Analysis and Sorting in Plant Genomics [J]. Chinese Bulletin of Botany, 2024, 59(5): 774-782. |
| [15] | QU Ze-Kun, ZHU Li-Qin, JIANG Qi, WANG Xiao-Hong, YAO Xiao-Dong, CAI Shi-Feng, LUO Su-Zhen, sCHEN Guang-Shui. Nutrient foraging strategies of arbuscular mycorrhizal tree species in a subtropical evergreen broadleaf forest and their relationship with fine root morphology [J]. Chin J Plant Ecol, 2024, 48(4): 416-427. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||