

Chinese Bulletin of Botany ›› 2016, Vol. 51 ›› Issue (2): 202-209.DOI: 10.11983/CBB15088 cstr: 32102.14.CBB15088
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Dongmei Li1, Luya Wang1, Lanyue Zhang2, Ziyang Tie2, Huiping Mao1,*( )
)
Received:2015-05-18
Accepted:2015-09-04
Online:2016-03-01
Published:2016-03-31
Contact:
E-mail: Dongmei Li, Luya Wang, Lanyue Zhang, Ziyang Tie, Huiping Mao. Mechanism of Arabidopsis Short Peptide Hormones PROPEP Gene Family in the Root Growth[J]. Chinese Bulletin of Botany, 2016, 51(2): 202-209.
| Primer name | Sequences (5'-3') | Function |
|---|---|---|
| AtPROPEP1_PF-HindIII AtPROPEP1_PR-BamHI AtPROPEP2_PF-HindIII AtPROPEP2_PR-BamHI AtPROPEP3_PF-BamHI AtPROPEP3_PR-NotI AtPROPEP4_PF-HindIII AtPROPEP4_PR-EcoRI AtPROPEP5_PF-HindIII AtPROPEP5_PR-BamHI AtPROPEP6_PF-XhoI AtPROPEP6_PR-EcoRI | CCCAAGCTTGTAAATTATAGTGAAAGGTACGG GCGGATCCTGAGATCTGATAAGACAGAGG CCCAAGCTTCGCATTCGCTTTTTTCTTTTTG GCGGATCCTGAAATCCAATAGTTTGTGAG GCGGATCCTATTTTAACAGTCAACAGCTATTTGG TTGCGGCCGCCGTTGACTTCTTAATCTTTTTTTG CCCAAGCTTAATAAGGATGAATAAAAAGTTTGGG CCGGAATTCGTTTTTCTTCAATTCTGCTTCGTG CCCAAGCTTTACTTAATTTCTTGTGAGAAACTTG GCGGATCCCTTCGCTATCTTCTAAGTTCCTC CCCTCGAGTGATATCTAAGTCCAACTTGGTG CCGGAATTCGTTTTTTGTTTTCTTTCTCTTCTT | AtPROPEP1 promoter clone AtPROPEP2 promoter clone AtPROPEP3 promoter clone AtPROPEP4 promoter clone AtPROPEP5 promoter clone AtPROPEP6 promoter clone |
Table 1 Sequences of AtPROPEPs promoter clone primers
| Primer name | Sequences (5'-3') | Function |
|---|---|---|
| AtPROPEP1_PF-HindIII AtPROPEP1_PR-BamHI AtPROPEP2_PF-HindIII AtPROPEP2_PR-BamHI AtPROPEP3_PF-BamHI AtPROPEP3_PR-NotI AtPROPEP4_PF-HindIII AtPROPEP4_PR-EcoRI AtPROPEP5_PF-HindIII AtPROPEP5_PR-BamHI AtPROPEP6_PF-XhoI AtPROPEP6_PR-EcoRI | CCCAAGCTTGTAAATTATAGTGAAAGGTACGG GCGGATCCTGAGATCTGATAAGACAGAGG CCCAAGCTTCGCATTCGCTTTTTTCTTTTTG GCGGATCCTGAAATCCAATAGTTTGTGAG GCGGATCCTATTTTAACAGTCAACAGCTATTTGG TTGCGGCCGCCGTTGACTTCTTAATCTTTTTTTG CCCAAGCTTAATAAGGATGAATAAAAAGTTTGGG CCGGAATTCGTTTTTCTTCAATTCTGCTTCGTG CCCAAGCTTTACTTAATTTCTTGTGAGAAACTTG GCGGATCCCTTCGCTATCTTCTAAGTTCCTC CCCTCGAGTGATATCTAAGTCCAACTTGGTG CCGGAATTCGTTTTTTGTTTTCTTTCTCTTCTT | AtPROPEP1 promoter clone AtPROPEP2 promoter clone AtPROPEP3 promoter clone AtPROPEP4 promoter clone AtPROPEP5 promoter clone AtPROPEP6 promoter clone |
| Primer name | Sequences (5'-3') | Function |
|---|---|---|
| AtPROPEP3_CDSF-SalI AtPROPEP3_CDSR-BamHI AtPROPEP4_CDSF-SalI AtPROPEP4_CDSR-EcoRI | GCGTCGACATGGAGAATCTCAGAAATGG CGGGATCCCTAATTGTGTTTGCCTCCTT GCGTCGACATGGAGAGAGGAGTTTCTTA CGGAATTCCTAAAACGGCTTCTTGTTGG | AtPROPEP3 coding sequence clone AtPROPEP4 coding sequence clone |
Table 2 Sequences of AtPROPEP3 and AtPROPEP4 clone primers
| Primer name | Sequences (5'-3') | Function |
|---|---|---|
| AtPROPEP3_CDSF-SalI AtPROPEP3_CDSR-BamHI AtPROPEP4_CDSF-SalI AtPROPEP4_CDSR-EcoRI | GCGTCGACATGGAGAATCTCAGAAATGG CGGGATCCCTAATTGTGTTTGCCTCCTT GCGTCGACATGGAGAGAGGAGTTTCTTA CGGAATTCCTAAAACGGCTTCTTGTTGG | AtPROPEP3 coding sequence clone AtPROPEP4 coding sequence clone |
| Primer name | Sequences (5'-3') |
|---|---|
| AtPROPEP2F AtPROPEP2R AtPROPEP3F AtPROPEP3R AtPROPEP4F AtPROPEP4R AtPROPEP5F AtPROPEP5R AtPROPEP6F AtPROPEP6R ACTINF ACTINR | CTCGACCAAGCTCTCATAGCTG CACAACGACATCATCGTCTTTC TCTTCTTCTTGCGATCTTTCGTCAT CTGAACTTGGCGTAGGCTTAGTC CTCAAGCTTCTCGGTTTGCGATC ACTTTCTCTCGACTTCTTTAGTAC GAGATTGTTGCAAGCTCATGCCTC AGTTGAAGTTTCGATAGATGAAGGT TGAAGTGTCTTGGTCTTGAGTC TGGTCTCCTTCTTAACACTGCTG ACGGTAACATTGTGCTCAGTGGTG CTTGGAGATCCACATCTGCTGGA |
Table 3 Primers used for quantitative RT-PCR
| Primer name | Sequences (5'-3') |
|---|---|
| AtPROPEP2F AtPROPEP2R AtPROPEP3F AtPROPEP3R AtPROPEP4F AtPROPEP4R AtPROPEP5F AtPROPEP5R AtPROPEP6F AtPROPEP6R ACTINF ACTINR | CTCGACCAAGCTCTCATAGCTG CACAACGACATCATCGTCTTTC TCTTCTTCTTGCGATCTTTCGTCAT CTGAACTTGGCGTAGGCTTAGTC CTCAAGCTTCTCGGTTTGCGATC ACTTTCTCTCGACTTCTTTAGTAC GAGATTGTTGCAAGCTCATGCCTC AGTTGAAGTTTCGATAGATGAAGGT TGAAGTGTCTTGGTCTTGAGTC TGGTCTCCTTCTTAACACTGCTG ACGGTAACATTGTGCTCAGTGGTG CTTGGAGATCCACATCTGCTGGA |
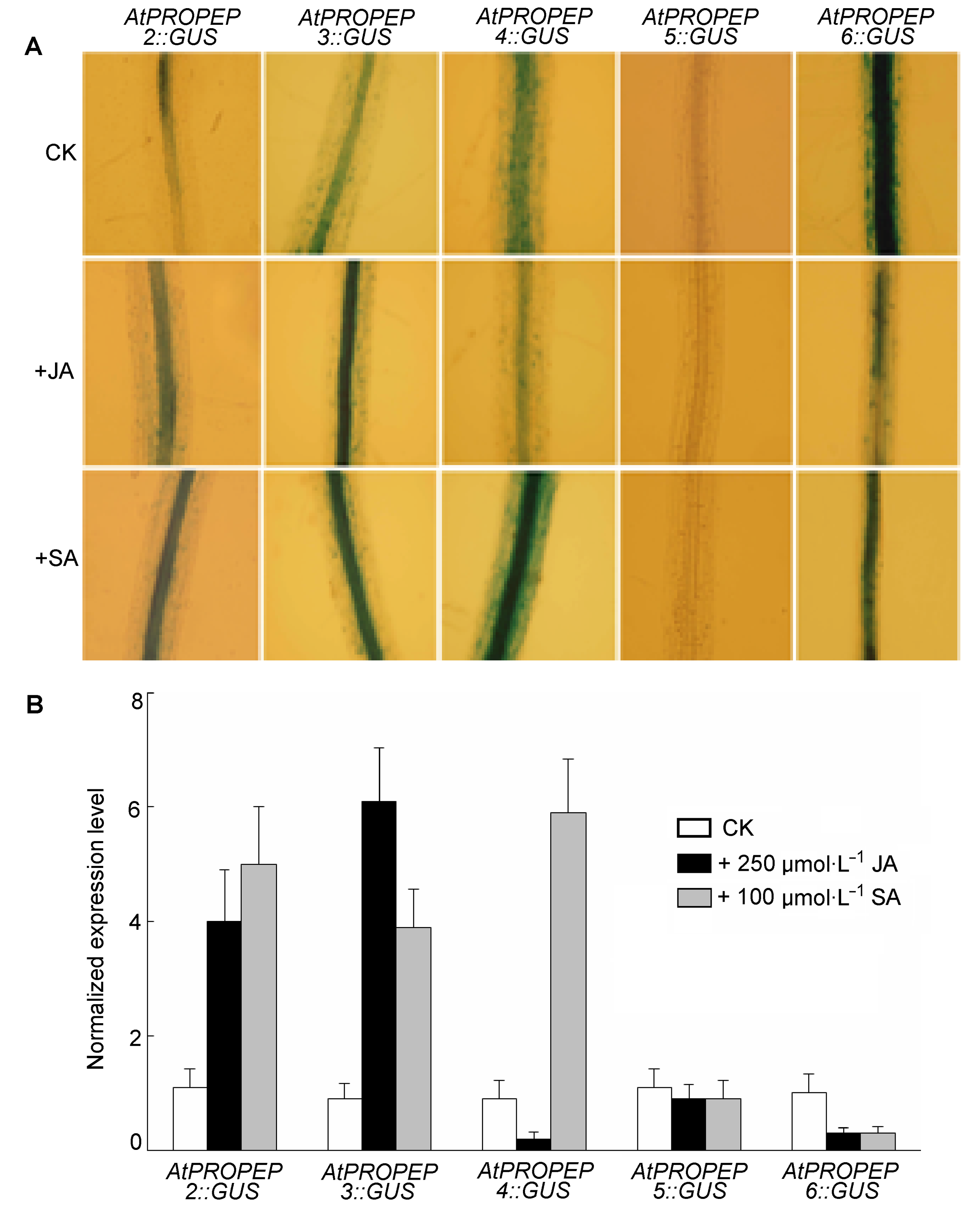
Figure 2 Response of AtPROPEP2,3−6 promoter to jasmonic acid (JA) and salicylic acid (SA) (A) GUS staining of transgenic seedlings harboring AtPROPEP2,3−6::GUS construct under 250 µmol∙L−1 JA and 100 µmol∙L−1 SA; (B) Quantitative RT-PCR analysis of AtPROPEP2,3−6 expression level under JA and SA treatments
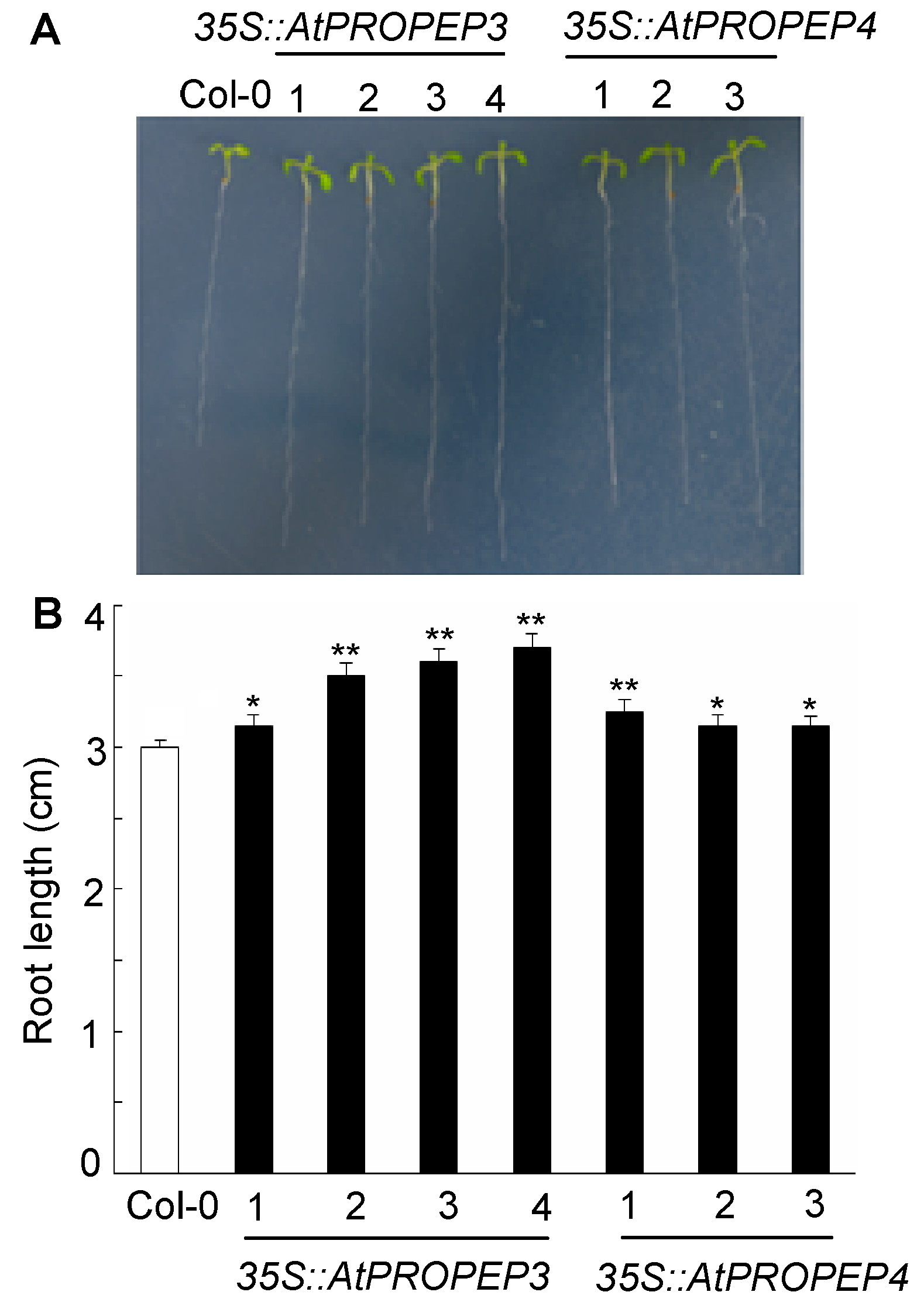
Figure 4 Arabidopsis plants overexpressing AtPROPEP3 and AtPROPEP4 exhibit longer root than that of wild type (A) Seedlings of Col-0 and transgenic seedlings grown on CK medium; (B) Measurement of root growth under CK medium (**P<0.01; * P<0.05)
| [1] | 李新锋, 赵淑清 (2004). 转基因植物中报告基因GUS的活性检测及其应用. 生命的化学 24, 71-74. |
| [2] | Bartels S, Lori M, Mbengue M, van Verk M, Klauser D, Hander T, Böni R, Robatzek S, Boller T (2013). The family of Peps and their precursors in Arabidopsis: dif- ferential expression and localization but similar induction of pattern-triggered immune responses. J Exp Bot 64, 5309-5321. |
| [3] | Gijrlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, Ryals J (1996). Bekothiadiazdle, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8, 629-643. |
| [4] |
Huffaker A, Pearce G, Ryan CA (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103, 10098-10103.
DOI PMID |
| [5] |
Huffaker A, Ryan CA (2007). Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA 104, 10732-10736.
DOI PMID |
| [6] | Kuc J (1982). Induced immunity to plant disease. Bioscience 32, 854-860. |
| [7] | Ma C, Guo J, Kang Y, Doman K, Bryan AC, Tax FE, Yamaguchi Y, Qi Z (2014). AtPEPTIDE RECEPTOR2 mediates the AtPEPTIDE1 induced cytosolic Ca2+ rise which is required for the suppression of Glutamate Dumper gene expression in Arabidopsis roots. J Integr Plant Biol 56, 684-694. |
| [8] | Pearce G, Moura DS, Stratmann J, Ryan CA (2001). Production of multiple plant hormones from a single polyprotein precursor. Nature 411, 817-820. |
| [9] |
Ross AF (1961). Systemic acquired resistance induced by localized virus infections in plants. Virology 14, 340-358.
PMID |
| [10] |
Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645-656.
DOI PMID |
| [11] | Van Loon LC, Van Kammen A (1970). Polyacrylamide disc electrophoresis of the soluble proteins from Nicotiene tabacum var. samsun and Samnkun NN. II. Changes in protein constitution after infection with tobacco mosaic virus. Virology 40, 199-211. |
| [12] |
Vijayan P, Shockey J, Lévesque CA, Cook RJ, Browse J (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc Natl Acad Sci USA 95, 7209-7214.
DOI PMID |
| [13] | Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA (1991). Co-ordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085-1094. |
| [14] | Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508-522. |
| [15] | Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip methods. Nature Protocols 1, 641-646. |
| [16] |
Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Some- rville S (2004). Host and non-host pathogens elicit differ- ent jasmonate/ethylene responses in Arabidopsis. Plant J 40, 633-646.
DOI PMID |
| [1] | ZHANG Zi-Rui, Zhou Jing, HU Yan-Ping, Liang Shuang, MA Yong-Peng, CHEN Wei-Le. Root-associated Fungal Communities of the Critically Endangered Plant Pinus Squamata [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | WANG Xiu-Yuan, SHEN Lei, LIU Ting-Ting, WEI Wen-Wen, ZHANG Shuai, ZHANG Wei. Spatial and temporal distribution of root system and interspecific competition strategy in Malus pumila ‘Saiwaihong’ - Glycine max agroforestry system [J]. Chin J Plant Ecol, 2025, 49(5): 748-759. |
| [3] | Can Ye, Linbo Yao, Ying Jin, Rong Gao, Qi Tan, Xuying Li, Yanjun Zhang, Xifeng Chen, Bojun Ma, Wei Zhang, Kewei Zhang. Establishment and Application of a High-throughput Screening Method for Salicylic Acid Metabolic Mutants in Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [4] | Yuhan Liu, Qijiang Cao, Shihan Zhang, Yihui Li, Jing Wang, Xiaomeng Tan, Xiaoru Liu, Xianling Wang. Mechanism of AtFTCD-L in Root Response to Soil Compaction [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [5] | LI Meng-Qi, MIAO Ling-Feng, LI Da-Dong, LONG Yi-Fan, YE Bing-Bing, YANG Fan. Response of mangrove fine root functional traits to sediment nutrient changes at different tide levels in Dongzhaigang, Hainan, China [J]. Chin J Plant Ecol, 2025, 49(4): 552-561. |
| [6] | WANG Juan, ZHANG Deng-Shan, XIAO Yuan-Ming, PEI Quan-Bang, WANG Bo, FAN Bo, ZHOU Guo-Ying. Relationships between the characteristics of root exudate and environmental factors in the alpine steppe following long-term grazing exclusion [J]. Chin J Plant Ecol, 2025, 49(4): 596-609. |
| [7] | GUO Li-Qi, YAN Xiao-Lei, CAO Lei, GAO Jing, LIU Rui-Qiang, ZHOU Xu-Hui. Effects of mycorrhizal types and root traits of tree species on rhizosphere microbial network complexity [J]. Chin J Plant Ecol, 2025, 49(4): 573-584. |
| [8] | DU Ying-Jie, FAN Ai-Lian, WANG Xue, YAN Xiao-Jun, CHEN Ting-Ting, JIA Lin-Qiao, JIANG Qi, CHEN Guang-Shui. Coordination and differences in root-leaf functional traits between tree species and understory shrub species in a subtropical natural evergreen broadleaf forest [J]. Chin J Plant Ecol, 2025, 49(4): 585-595. |
| [9] | Zeng Wendan, Yan Huabing, Wu Zhengdan, Shang Xiaohong, Cao Sheng, Lu Liuying, Xiao Liang, Shi Pingli, Cheng Dong, Long Ziyuan, Li Jieyu. Agrobacterium rhizogenes-mediated Transformation System of Pueraria lobata Hairy Roots [J]. Chinese Bulletin of Botany, 2025, 60(3): 425-434. |
| [10] | Yang Li, Qu Xitong, Chen Zihang, Zou Tingting, Wang Quanhua, Wang Xiaoli. Identification of the Spinach AT-hook Gene Family and Analysis of Expression Profiles [J]. Chinese Bulletin of Botany, 2025, 60(3): 377-392. |
| [11] | Gu Jingjing, Liu Yizhuo, Su Yang. The functions and challenges of grass-roots local governments in fulfilling the Kunming-Montreal Global Biodiversity Framework—A comparative analysis with the objectives of the United Nations Framework Convention on Climate Change [J]. Biodiv Sci, 2025, 33(3): 24585-. |
| [12] | ZHENG Lin-Min, XIONG Xiao-Ling, JIANG Yong-Meng, WANG Man, ZHANG Jin-Xiu, ZENG Zhi-Wei, LYU Mao-Kui, XIE Jin-Sheng. Decomposition regularities of leaf litter and fine roots of Cunninghamia lanceolata and their divergent drivers at different altitudes in the Wuyi Mountain [J]. Chin J Plant Ecol, 2025, 49(2): 244-255. |
| [13] | NIU Yun-Ming, JIA Guo-Dong, WANG Xin, LIU Zi-He. Dynamic changes of transpiration water age and water utilization strategies for trees at different altitudes in Lushan area [J]. Chin J Plant Ecol, 2024, 48(9): 1104-1117. |
| [14] | PENG Si-Rui, ZHANG Hui-Ling, SUN Zhao-Lin, ZHAO Xue-Chao, TIAN Peng, CHEN Di-Ma, WANG Qing-Kui, LIU Sheng-En. Effects of long-term litter removal on soil organic carbon and multiple components in subtropical Cunninghamia lanceolata forest [J]. Chin J Plant Ecol, 2024, 48(8): 1078-1088. |
| [15] | LONG Ji-Lan, JIANG Zheng, LIU Ding-Qin, MIAO Yu-Xuan, ZHOU Ling-Yan, FENG Ying, PEI Jia-Ning, LIU Rui-Qiang, ZHOU Xu-Hui, FU Yu-Ling. Effects of drought on plant root exudates and associated rhizosphere priming effect: review and prospect [J]. Chin J Plant Ecol, 2024, 48(7): 817-827. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||