

Chinese Bulletin of Botany ›› 2020, Vol. 55 ›› Issue (4): 457-467.DOI: 10.11983/CBB19228 cstr: 32102.14.CBB19228
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Jiuxiang Huang1,Wenna Chen2,Yuling Li1,Gang Yao1,*( )
)
Received:2019-11-23
Accepted:2020-03-01
Online:2020-07-01
Published:2020-05-21
Contact:
Gang Yao
Jiuxiang Huang,Wenna Chen,Yuling Li,Gang Yao. Phylogenetic Study of Amaranthaceae sensu lato Based on Multiple Plastid DNA Fragments[J]. Chinese Bulletin of Botany, 2020, 55(4): 457-467.
| Name of DNA sequences | Length of DNA sequences (bp) | Model selected |
|---|---|---|
| atpB | 1497 | GTR+I+Γ |
| ndhF | 2242 | TVM+I+Γ |
| psbB | 1527 | GTR+I+Γ |
| rbcL | 1344 | TVM+I+Γ |
| rpoC2 | 4147 | GTR+I+Γ |
| rps4 | 606 | TVM+Γ |
| rps16 | 1401 | GTR+I+Γ |
| matK/trnK | 2757 | GTR+I+Γ |
Table 1 Information of DNA sequences used in the study
| Name of DNA sequences | Length of DNA sequences (bp) | Model selected |
|---|---|---|
| atpB | 1497 | GTR+I+Γ |
| ndhF | 2242 | TVM+I+Γ |
| psbB | 1527 | GTR+I+Γ |
| rbcL | 1344 | TVM+I+Γ |
| rpoC2 | 4147 | GTR+I+Γ |
| rps4 | 606 | TVM+Γ |
| rps16 | 1401 | GTR+I+Γ |
| matK/trnK | 2757 | GTR+I+Γ |
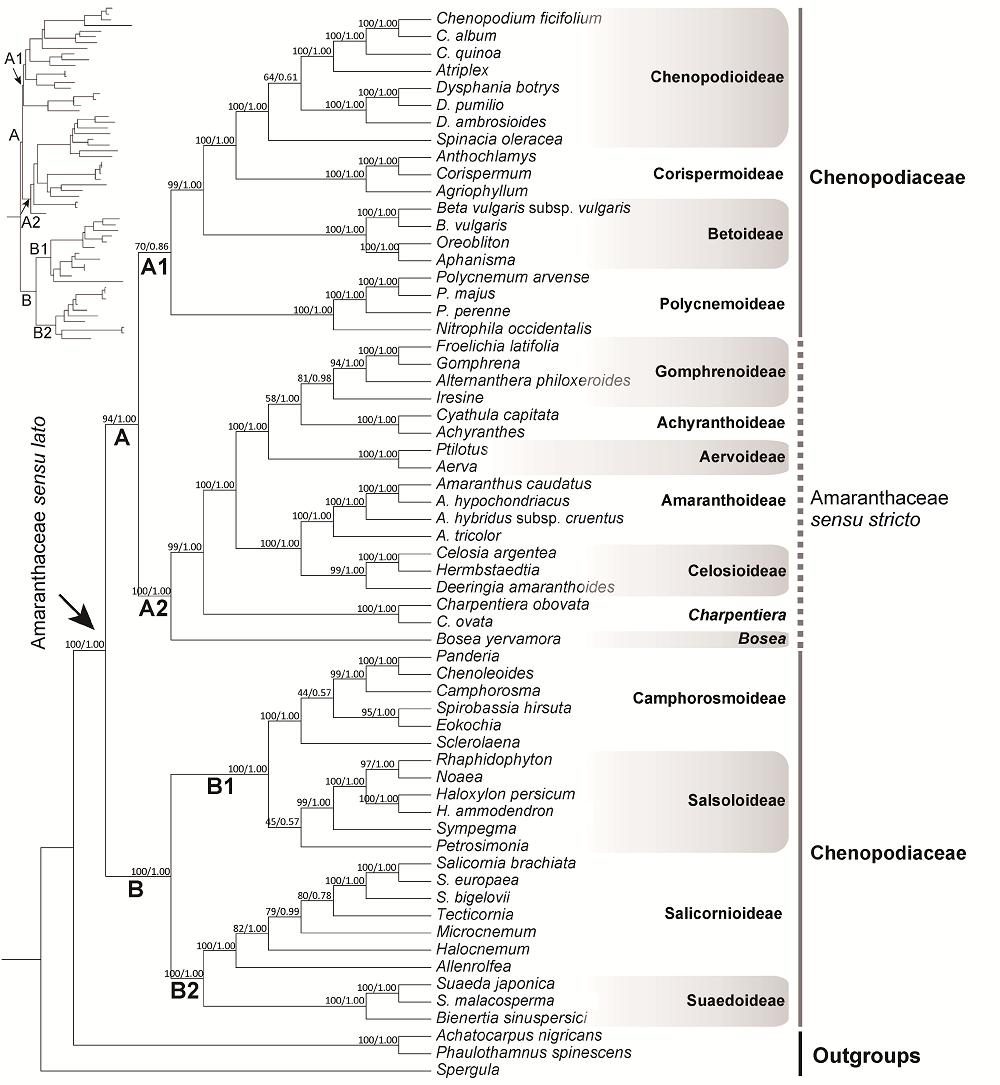
Figure 1 Phylogeny of Amaranthaceae sensu lato inferred from the combined matrix of eight plastid DNA regions Bootstrap support value and posterior probability of each node are indicated above branches.
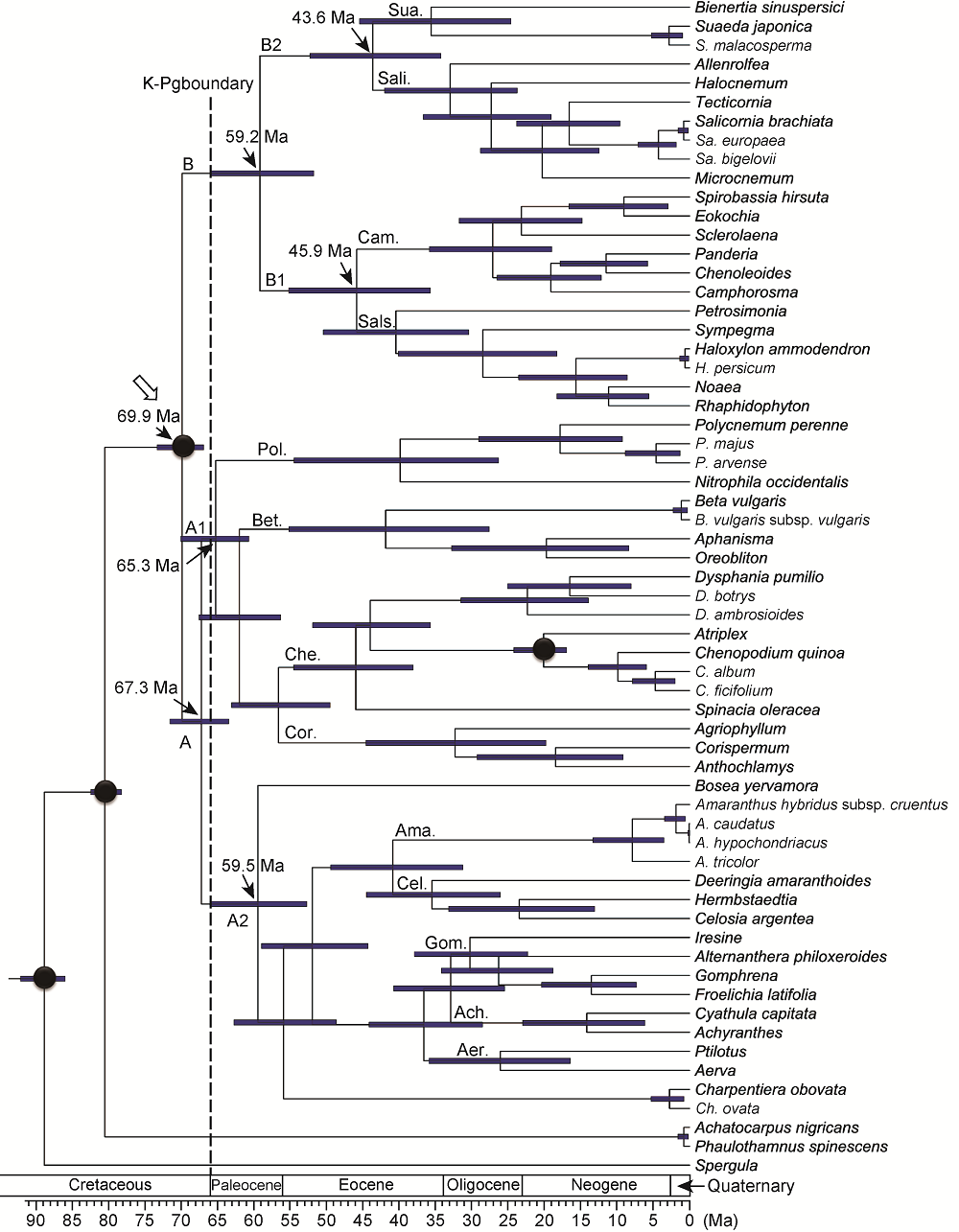
Figure 2 Chronogram based on the BEAST analysis of the combined matrix of eight plastid DNA regions and four calibrations Calibration points are depicted with black circles. The crown node of Amaranthaceae sensu lato is shown by a hollow arrowhead. Ach: Achyranthoideae; Aer: Aervoideae; Ama: Amaranthoideae; Bet: Betoideae; Cam: Camphorosmoideae; Cel: Celosioideae; Che: Chenopodioideae; Cor: Corispermoideae; Gom: Gomphrenoideae; Pol: Polycnemoideae; Sali: Salicornioideae; Sals: Salsoloideae; Sua: Suaedoideae
| [1] | 中国科学院中国植物志编辑委员会 (1979). 中国植物志, 第25卷第2分册. 北京: 科学出版社. pp. 241. |
| [2] | APG IV (2016). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181, 1-20. |
| [3] | Bentham G, Hooker JD (1880). Genera Plantarum, Vol. 3. London: Lovell Reeve. pp. 1258. |
| [4] | Brockington SF, Alexandre R, Ramdial J, Moore MJ, Crawley S, Dhingra A, Hilu K, Soltis DE, Soltis PS (2009). Phylogeny of the Caryophyllales sensu lato: revisiting hypotheses on pollination biology and perianth differentiation in the core Caryophyllales. Int J Plant Sci 170, 627-643. |
| [5] | Christenhusz MJM, Byng JW (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261, 201-217. |
| [6] | Cronquist A (1988). Caryophyllidae. In: The Evolution and Classification of Flowering Plants, 2nd edn. New York: The New York Botanical Garden. pp. 309-320. |
| [7] | Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002). Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am J Bot 89, 132-144. |
| [8] | Drummond AJ, Suchard MA, Xie D, Rambaut A (2012). Bayesian phylogenetics with BEAUTi and the BEAST 1.7. Mol Biol Evol 29, 1967-1973. |
| [9] | Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJD, Clements MA, Arroyo MTK, Leebens-Mack J, Endara L, Kriebel R, Neubig KM, Whitten WM, Williams NH, Cameron KM (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc R Soc B Biol Sci 282, 20151553. |
| [10] | Guo X, Thomas DC, Saunders RMK (2018). Gene tree discordance and coalescent methods support ancient intergeneric hybridisation between Dasymaschalon and Friesodielsia(Annonaceae). Mol Phylogenet Evol 127, 14-29. |
| [11] | Gurushidze M, Fritsch RM, Blattner FR (2010). Species-level phylogeny of Allium subgenus Melanocrommyum: incomplete lineage sorting, hybridization and trnF gene duplication. Taxon 59, 829-840. |
| [12] | Hernández-Ledesma P, Berendsohn WG, Borsch T, von Mering S, Akhani H, Arias S, Castañeda-Noa I, Eggli U, Eriksson R, Flores-Olvera H, Fuentes-Bazán S, Kadereit G, Klak C, Korotkova N, Nyffeler R, Ocampo G, Ochoterena H, Oxelman B, Rabeler RK, Sanchez A, Schlumpberger BO, Uotila P (2015). A taxonomic backbone for the global synthesis of species diversity in the angiosperm order Caryophyllales. Willdenowia 45, 281-383. |
| [13] | Kadereit G, Ackerly D, Pirie MD (2012). A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proc R Soc B Biol Sci 279, 3304-3311. |
| [14] |
Kadereit G, Borsch T, Weising K, Freitag H (2003). Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. Int J Plant Sci 164, 959-986.
DOI URL |
| [15] | Kadereit G, Gotzek D, Jacobs S, Freitag H (2005). Origin and age of Australian Chenopodiaceae. Org Divers Evol 5, 59-80. |
| [16] |
Katoh K, Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772-780.
URL PMID |
| [17] | Koenen EJM, Clarkson JJ, Pennington TD, Chatrou LW (2015). Recently evolved diversity and convergent radiations of rainforest mahoganies (Meliaceae) shed new light on the origins of rainforest hyperdiversity. New Phytol 207, 327-339. |
| [18] | Kühn U, Bittrich V, Carolin R, Freitag H, Hedge IC, Uotila P, Wilson PG (1993). Chenopodiaceae. In: Kubitzki K, ed. Families and Genera of Vascular Plants, Vol. 2. Berlin: Springer. pp. 253-281. |
| [19] |
Li HT, Yi TS, Gao LM, Ma PF, Zhang T, Yang JB, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, Wang H, van der Bank M, Zhang SD, Wang QF, Wang J, Zhang ZR, Fu CN, Yang J, Hollingsworth PM, Chase MW, Soltis DE, Soltis PS, Li DZ (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat Plants 5, 461-470.
URL PMID |
| [20] |
Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T (2015). A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol 207, 437-453.
URL PMID |
| [21] | Masson R, Kadereit G (2013). Phylogeny of Polycnemoideae (Amaranthaceae): implications for biogeography, character evolution and taxonomy. Taxon 62, 100-111. |
| [22] | Miller MA, Pfeiffer W, Schwartz T (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA: IEEE. pp. 1-8. |
| [23] | Moquin-Tandon A (1837). Note sur le genre Polycnemum et sur une nouvelle tribu de la famille des Paronychees. Ann Sci Nat 7, 33-42. |
| [24] | Muller J (1981). Fossil pollen records of extant angiosperms. Bot Rev 47, 1-142. |
| [25] | Müler K, Borsch T (2005). Phylogenetics of Amaranthaceae based on matK/trnK sequence data—evidence from parsimony, likelihood, and Bayesian analyses. Ann Missouri Bot Gard 92, 66-102. |
| [26] |
Posada D (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253-1256.
DOI URL PMID |
| [27] | Pratt DB (2003). Phylogeny and Morphological Evolution of the Chenopodiaceae-Amaranthaceae Alliance. Ph.D. thesis. Ames: Iowa State University. pp. 116. |
| [28] | Rambaut A (2012). FigTree version 1.4.0. Available from: http://tree.bio.ed.ac.uk/software/figtree/. |
| [29] | Rambaut A, Suchard MA, Drummond AJ (2014). Tracer v1.6. Available from: http://beast.bio.ed.ac.uk/Tracer/. |
| [30] |
Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574.
URL PMID |
| [31] |
Schulte P, Alegret L, Arenillas I, Arz JA, Barton PJ, Bown PR, Bralower TJ, Christeson GL, Claeys P, Cockell CS, Collins GS, Deutsch A, Goldin TJ, Goto K, Grajales-Nishimura JM, Grieve RAF, Gulick SPS, Johnson KR, Kiessling W, Koeberl C, Kring DA, MacLeod KG, Matsui T, Melosh J, Montanari A, Morgan JV, Neal CR, Nichols DJ, Norris RD, Pierazzo E, Ravizza G, Rebolledo-Vieyra M, Reimold WU, Robin E, Salge T, Speijer RP, Sweet AR, Urrutia-Fucugauchi J, Vajda V, Whalen MT, Willumsen PS (2010). The chicxulub asteroid impact and mass extinction at the Cretaceous-Paleogene boundary. Science 327, 1214-1218.
DOI URL PMID |
| [32] | Scott AJ (1977a). Nomina conservanda proposita. Taxon 26, 246. |
| [33] | Scott AJ (1977b). Reinstatement and revision of Salicorniaceae J. Agardh (Caryophyllales). Bot J Linn Soc 75, 357-374. |
| [34] |
Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, Brockington SF, Refulio-Rodriguez NF, Walker JB, Moore MJ, Carlsward BS, Bell CD, Latvis M, Crawley S, Black C, Diouf D, Xi ZX, Rushworth CA, Gitzendanner MA, Sytsma KJ, Qiu YL, Hilu KW, Davis CC, Sanderson MJ, Beaman RS, Olmstead RG, Judd WS, Donoghue MJ, Soltis PS (2011). Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot 98, 704-730.
URL PMID |
| [35] | Soriano A (1944). El género Nitrophila en la Argentina y su posiciónsistemática. Rev Argent Agron 11, 302. |
| [36] |
Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood- based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688-2690.
DOI URL PMID |
| [37] | Stevens PF (2001). Angiosperm Phylogeny Website. Version 14, July 2017 [and more or less continuously updated since]. Available from: http://www.mobot.org/MOBOT/research/APweb/. |
| [38] |
Sukhorukov AP, Mavrodiev EV, Struwig M, Nilova MV, Dzhalilova KK, Balandin SA, Erst A, Krinitsyna AA (2015). One-seeded fruits in the core Caryophyllales: their origin and structural diversity. PLoS One 10, e0117974.
URL PMID |
| [39] | Takhtajan A (1997). Diversity and Classification of Flowering Plants. New York: Columbia University Press. pp. 1-643. |
| [40] | Ulbrich E (1934). Chenopodiaceae. In: Engler A, Prantl K, eds. Die Naturlichen Pflanzenfamilien, Vol. 16c. Leipzig: Engelmann. pp. 379-584. |
| [41] |
Walker JF, Yang Y, Feng T, Timoneda A, Mikenas J, Hutchison V, Edwards C, Wang N, Ahluwalia S, Olivieri J, Walker-Hale N, Majure LC, Puente R, Kadereit G, Lauterbach M, Eggli U, Flores-Olvera H, Ochoterena H, Brockington SF, Moore MJ, Smith SA (2018). From cacti to carnivores: improved phylotranscriptomic sampling and hierarchical homology inference provide further insight into the evolution of Caryophyllales. Am J Bot 105, 446-462.
DOI URL PMID |
| [42] | Wang W, Ortiz RDC, Jacques FMB, Xiang XG, Li HL, Lin L, Li RQ, Liu Y, Soltis PS, Soltis DE, Chen ZD (2012). Menispermaceae and the diversification of tropical rainforests near the Cretaceous-Paleogene boundary. New Phytol 195, 470-478. |
| [43] | Wikström N, Savolainen V, Chase MW (2001). Evolution of the angiosperms: calibrating the family tree. Proc R Soc B Biol Sci 268, 2211-2220. |
| [44] |
Yang Y, Moore MJ, Brockington SF, Mikenas J, Olivieri J, Walker JF, Smith SA (2018). Improved transcriptome sampling pinpoints 26 ancient and more recent polyploidy events in Caryophyllales, including two allopolyploidy events. New Phytol 217, 855-870.
DOI URL PMID |
| [45] |
Yao G, Jin JJ, Li HT, Yang JB, Mandala VS, Croley M, Mostow R, Douglas NA, Chase MW, Christenhusz MJM, Soltis DE, Soltis PS, Smith SA, Brockington SF, Moore MJ, Yi TS, Li DZ (2019). Plastid phylogenomic insights into the evolution of Caryophyllales. Mol Phylogenet Evol 134, 74-86.
DOI URL PMID |
| [46] |
Yi TS, Jin GH, Wen J (2015). Chloroplast capture and intra- and inter-continental biogeographic diversification in the Asian—New World disjunct plant genus Osmorhiza(Apiaceae). Mol Phylogenet Evol 85, 10-21.
URL PMID |
| [1] | Hong Deyuan. A brief discussion on methodology in taxonomy [J]. Biodiv Sci, 2025, 33(2): 24541-. |
| [2] | Yajun Sun. What do higher or lower organisms mean—Clarify the meaning and validity of the biological ladder implied by On the Origin of Species [J]. Biodiv Sci, 2025, 33(1): 24394-. |
| [3] | Hua He, Dunyan Tan, Xiaochen Yang. Cryptic dioecy in angiosperms: Diversity, phylogeny and evolutionary significance [J]. Biodiv Sci, 2024, 32(6): 24149-. |
| [4] | Yanyu Ai, Haixia Hu, Ting Shen, Yuxuan Mo, Jinhua Qi, Liang Song. Vascular epiphyte diversity and the correlation analysis with host tree characteristics: A case in a mid-mountain moist evergreen broad-leaved forest, Ailao Mountains [J]. Biodiv Sci, 2024, 32(5): 24072-. |
| [5] | Yanwen Lv, Ziyun Wang, Yu Xiao, Zihan He, Chao Wu, Xinsheng Hu. Advances in lineage sorting theories and their detection methods [J]. Biodiv Sci, 2024, 32(4): 23400-. |
| [6] | Zhi Yang, Yong Yang. Research Advances on Nuclear Genomes of Economically Important Trees of Lauraceae [J]. Chinese Bulletin of Botany, 2024, 59(2): 302-318. |
| [7] | Chen-Kun Jiang, Wen-Bin Yu, Guang-Yuan Rao, Huaicheng Li, Julien B. Bachelier, Hartmut H. Hilger, Theodor C. H. Cole. Plant Phylogeny Posters—An educational project on plant diversity from an evolutionary perspective [J]. Biodiv Sci, 2024, 32(11): 24210-. |
| [8] | Zhenzhou Chu, Gulbar Yisilam, Zezhong Qu, Xinmin Tian. Comparative Analyses on the Chloroplast Genome of Three Sympatric Atraphaxis Species [J]. Chinese Bulletin of Botany, 2023, 58(3): 417-432. |
| [9] | Zhizhong Li, Shuai Peng, Qingfeng Wang, Wei Li, Shichu Liang, Jinming Chen. Cryptic diversity of the genus Ottelia in China [J]. Biodiv Sci, 2023, 31(2): 22394-. |
| [10] | Jinbo Bao, Zhijie Ding, Haoyu Miao, Xueli Li, Shuxian Ren, Ruoyan Jiao, Hao Li, Qianqian Deng, Yingzi Li, Xinmin Tian. Analysis of Chloroplast Genomes of Aleurites moluccana [J]. Chinese Bulletin of Botany, 2023, 58(2): 248-260. |
| [11] | Huiyin Song, Zhengyu Hu, Guoxiang Liu. Assessing advances in taxonomic research on Chlorellaceae (Chlorophyta) [J]. Biodiv Sci, 2023, 31(2): 22083-. |
| [12] | Ting Wang, Jiangping Shu, Yufeng Gu, Yanqing Li, Tuo Yang, Zhoufeng Xu, Jianying Xiang, Xianchun Zhang, Yuehong Yan. Insight into the studies on diversity of lycophytes and ferns in China [J]. Biodiv Sci, 2022, 30(7): 22381-. |
| [13] | Jianming Wang, Mengjun Qu, Yin Wang, Yiming Feng, Bo Wu, Qi Lu, Nianpeng He, Jingwen Li. The drivers of plant taxonomic, functional, and phylogenetic β-diversity in the gobi desert of northern Qinghai-Tibet Plateau [J]. Biodiv Sci, 2022, 30(6): 21503-. |
| [14] | XIANG Wei, HUANG Dong-Liu, ZHU Shi-Dan. Absorptive root anatomical traits of 26 tropical and subtropical fern species [J]. Chin J Plant Ecol, 2022, 46(5): 593-601. |
| [15] | Fanbin Hu, Yue Xin, Ke Guo, Liqing Zhao. Seven newly recorded species from Xizang and Xinjiang in China [J]. Biodiv Sci, 2021, 29(9): 1265-1270. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||