

Chinese Bulletin of Botany ›› 2019, Vol. 54 ›› Issue (1): 23-36.DOI: 10.11983/CBB18064 cstr: 32102.14.CBB18064
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Xiaoxi Zhen,Haoran Liu,Xin Li,Fan Xu( ),Wenzhong Zhang(
),Wenzhong Zhang( )
)
Received:2018-03-12
Accepted:2018-07-16
Online:2019-01-01
Published:2019-07-31
Contact:
Fan Xu,Wenzhong Zhang
Xiaoxi Zhen,Haoran Liu,Xin Li,Fan Xu,Wenzhong Zhang. Heterologous Overexpression of Autophagy-related Gene OsATG8b from Rice Confers Tolerance to Nitrogen/Carbon Starvation and Increases Yield in Arabidopsis[J]. Chinese Bulletin of Botany, 2019, 54(1): 23-36.
| Primer name | Sequence (5′-3′) | Function |
|---|---|---|
| cOsATG8b-F | CCATTCAAGTGGATGGCCAAGAGCTCGTTCAAGC | Gene cloning |
| cOsATG8b-R | GGTGACCTAGAGCAGCCCAAAGGTGTTCTCG | Gene cloning |
| cpOsATG8b-F | AAGCTTAAAATTAAATAAGACGAACAGTCAAACG | Gene cloning |
| cpOsATG8b-R | CCATGGCGCTCCTTCCTGCACACAAT | Gene cloning |
| rtOsATG8b-F | GCTGATCTTACCGTTGGGCA | Real-time RT-PCR |
| rtOsATG8b-R | ATCAGAGCAGCTGTTGGTGG | Real-time RT-PCR |
| rtAtAMT1-F | GCCTCTGCTGACTACTCCAACTT | Real-time RT-PCR |
| rtAtAMT1-R | GACCAGAACCAGTGAGAGACGA | Real-time RT-PCR |
| rtAtNR1-F | AGGATGGGCTAGTAAGCATAAGG | Real-time RT-PCR |
| rtAtNR1-R | GCAAACTGAATCATAGGCGGTG | Real-time RT-PCR |
| rtAtGS2-F | CACCAAACCTTACTCTCTGACA | Real-time RT-PCR |
| rtAtGS2-R | CACTATCTTCACCAGGTGCTTG | Real-time RT-PCR |
| rtAtGDH1-F | GCTTTAGCAGCAACAAACAGAA | Real-time RT-PCR |
| rtAtGDH1-R | TGAGCCAATGCGTTCACTTC | Real-time RT-PCR |
| rtACTIN1-F | ACCATTGGTGCTGAGCGTTT | Real-time RT-PCR |
| rtACTIN1-R | CGCAGCTTCCATTCCTATGAA | Real-time RT-PCR |
| rtTIP41-F | GTATGAAGATGAACTGGCTGACAAT | Real-time RT-PCR |
| rtTIP41-R | ATCAACTCTCAGCCAAAATCGCAAG | Real-time RT-PCR |
Table 1 The information of primers
| Primer name | Sequence (5′-3′) | Function |
|---|---|---|
| cOsATG8b-F | CCATTCAAGTGGATGGCCAAGAGCTCGTTCAAGC | Gene cloning |
| cOsATG8b-R | GGTGACCTAGAGCAGCCCAAAGGTGTTCTCG | Gene cloning |
| cpOsATG8b-F | AAGCTTAAAATTAAATAAGACGAACAGTCAAACG | Gene cloning |
| cpOsATG8b-R | CCATGGCGCTCCTTCCTGCACACAAT | Gene cloning |
| rtOsATG8b-F | GCTGATCTTACCGTTGGGCA | Real-time RT-PCR |
| rtOsATG8b-R | ATCAGAGCAGCTGTTGGTGG | Real-time RT-PCR |
| rtAtAMT1-F | GCCTCTGCTGACTACTCCAACTT | Real-time RT-PCR |
| rtAtAMT1-R | GACCAGAACCAGTGAGAGACGA | Real-time RT-PCR |
| rtAtNR1-F | AGGATGGGCTAGTAAGCATAAGG | Real-time RT-PCR |
| rtAtNR1-R | GCAAACTGAATCATAGGCGGTG | Real-time RT-PCR |
| rtAtGS2-F | CACCAAACCTTACTCTCTGACA | Real-time RT-PCR |
| rtAtGS2-R | CACTATCTTCACCAGGTGCTTG | Real-time RT-PCR |
| rtAtGDH1-F | GCTTTAGCAGCAACAAACAGAA | Real-time RT-PCR |
| rtAtGDH1-R | TGAGCCAATGCGTTCACTTC | Real-time RT-PCR |
| rtACTIN1-F | ACCATTGGTGCTGAGCGTTT | Real-time RT-PCR |
| rtACTIN1-R | CGCAGCTTCCATTCCTATGAA | Real-time RT-PCR |
| rtTIP41-F | GTATGAAGATGAACTGGCTGACAAT | Real-time RT-PCR |
| rtTIP41-R | ATCAACTCTCAGCCAAAATCGCAAG | Real-time RT-PCR |
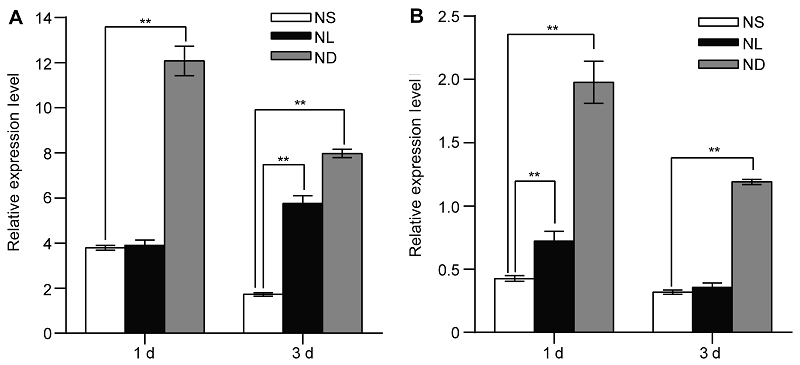
Figure 1 Identification of OsATG8b as a nitrogen deficiency inducible/responsive gene in leaves and roots of rice seedlings (A) The rice seedlings cultured with N-sufficient (NS) solution for 14 days and transferred to the same NS solution, low N (NL) solution and the N-deficient (ND) solution, the expression of OsATG8b gene in leaves after 1 day and 3 days treatment; (B) The expression of OsATG8b gene in roots after 1 day and 3 days treatment. Values are means±SD, n=16, three biological replicates were performed. ** indicate significant differences in NS solution compared with NL and ND solution (P<0.01) (Student’s t-test).
| WT | L-13 | L-14 | |
|---|---|---|---|
| Bloting time (d) | 36.56±1.58 | 30.78±2.07** | 31.39±1.91** |
| Flowering time (d) | 42.67±1.75 | 35.94±1.98** | 36.61±1.94** |
Table 2 Bolting and flowering times of the wild-type and 35S-OsATG8b transgenic Arabidopsis
| WT | L-13 | L-14 | |
|---|---|---|---|
| Bloting time (d) | 36.56±1.58 | 30.78±2.07** | 31.39±1.91** |
| Flowering time (d) | 42.67±1.75 | 35.94±1.98** | 36.61±1.94** |
| Total number of siliques | Yield per plant (mg) | Thousand grain weight (mg) | |
|---|---|---|---|
| WT | 35.74±3.86 | 85.34±7.89 | 14.87±0.23 |
| L-13 | 46.26±3.13** | 100.13±6.02** | 16.36±0.21** |
| L-14 | 48.22±3.62** | 99.77±5.76** | 17.54±0.41** |
Table 3 Yield related characteristics of the wild-type and 35S-OsATG8b transgenic Arabidopsis
| Total number of siliques | Yield per plant (mg) | Thousand grain weight (mg) | |
|---|---|---|---|
| WT | 35.74±3.86 | 85.34±7.89 | 14.87±0.23 |
| L-13 | 46.26±3.13** | 100.13±6.02** | 16.36±0.21** |
| L-14 | 48.22±3.62** | 99.77±5.76** | 17.54±0.41** |
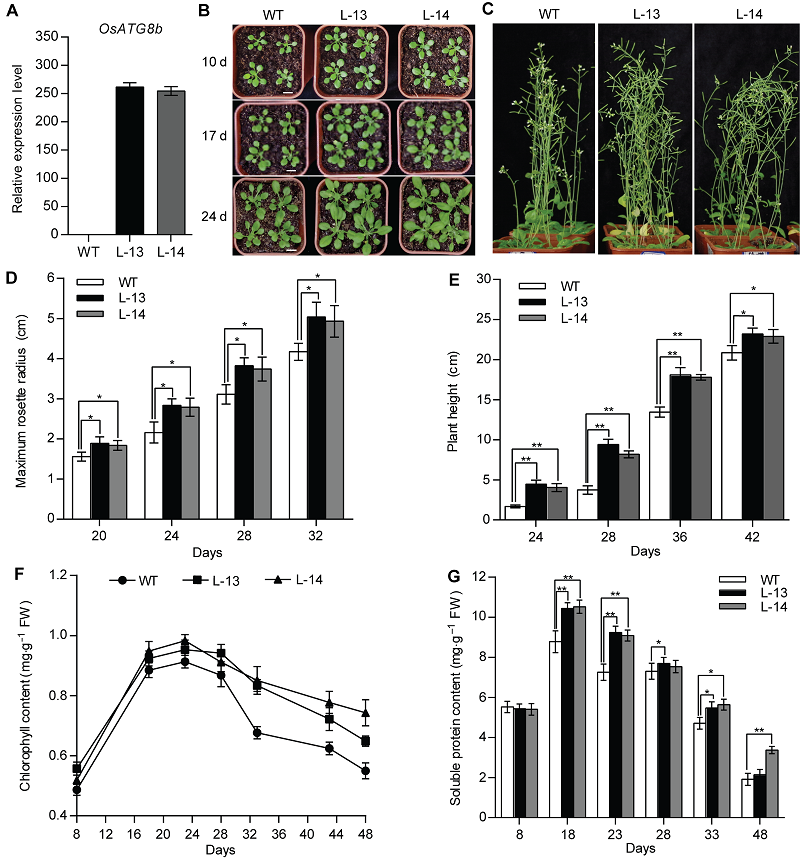
Figure 2 Over-expression of OsATG8b promotes growth and development of transgenic Arabidopsis 8-day-old seedlings were transferred to vermiculite-nutritional soil (1:3, v/v). (A) Expression level of OsATG8b in 14-day-old seedlings of 35S-OsATG8b transgenic lines and wild type (WT); (B) Panels from top to bottom show phenotypic observations of transgenic lines and WT of Arabidopsis at 10, 17 and 24 days after transfer to soil, respectively; (C) Phenotype of transgenic lines and WT at 42 days after transfer to soil; (D) The maximum rosette radius of 35S-OsATG8b transgenic lines and WT at different seedling age; (E) The plant height; (F) The total chlorophyll content; (G) The soluble protein content. Days: Days after germination. Values are means±SD, n=24, * P<0.05, ** P<0.01 (Student’s t-test), three biological replicates were performed. Bars=1 cm
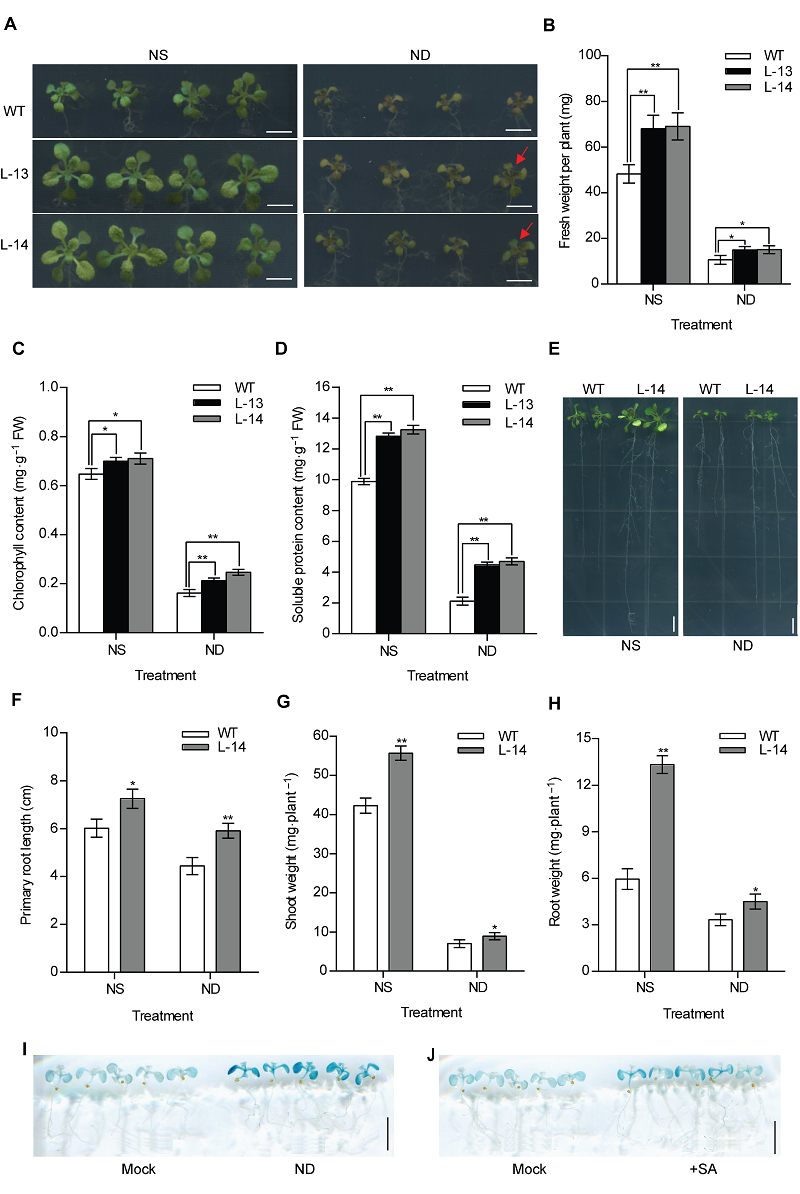
Figure 3 Overexpression of OsATG8b enhances tolerance to N deficiency in transgenic Arabidopsis (A) 7-day-old seedlings of 35S-OsATG8b transgenic lines and wild type (WT) were transferred to 1/2MS medium for horizontal culture with sufficient (NS) or deficient (ND) N for 9 days. (B)-(D) The fresh weight, chlorophyll content and soluble protein content in rosette leaves of WT and 35S-OsATG8b transgenic Arabidopsis under NS or ND for 9 days, respectively; (E) The phenotype of 7-day-old seedlings of 35S-OsATG8b transgenic lines and WT were transferred to vertical plates with NS or ND for 9 days; (F)-(H) The primary root length, the shoot weight and the root weight of WT and transgenic Arabidopsis lines under NS or ND for 9 days, respectively; (I), (J) 10-day-old seedlings of ProOsATG8b-GUS transgenic Arabidopsis were transferred to ND and 10 μmol·L-1 SA for 24 h, respectively. Mock represented that the seedlings without treated. Values are means±SD, n=16, * and ** indicate significant (P<0.05) and extremely significant (P<0.01) differences between transgenic lines and WT (Student’s t-test), three biological replicates were performed. Bars=5 mm
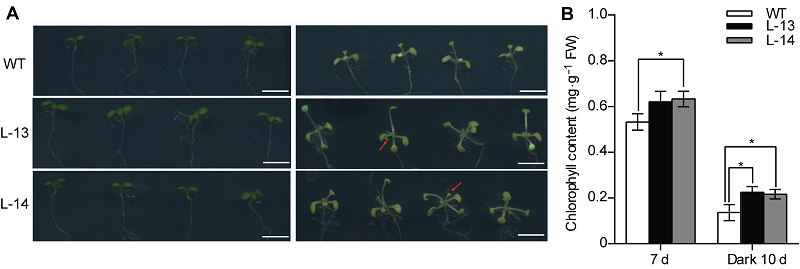
Figure 4 Overexpression of OsATG8b in Arabidopsis enhanced tolerance to carbon starvation induced by dark treatment (A) 7-day-old seedlings of 35S-OsATG8b transgenic lines and wild type (WT) were transferred to darkness for 10 days (The left is before treatment, and the right is after treatment); (B) The chlorophyll content determination. Values are means±SD, n=10, * indicate significant difference between transgenic lines and WT (P<0.05) (Student’s t-test), three biological replicates were performed. Bars=5 mm
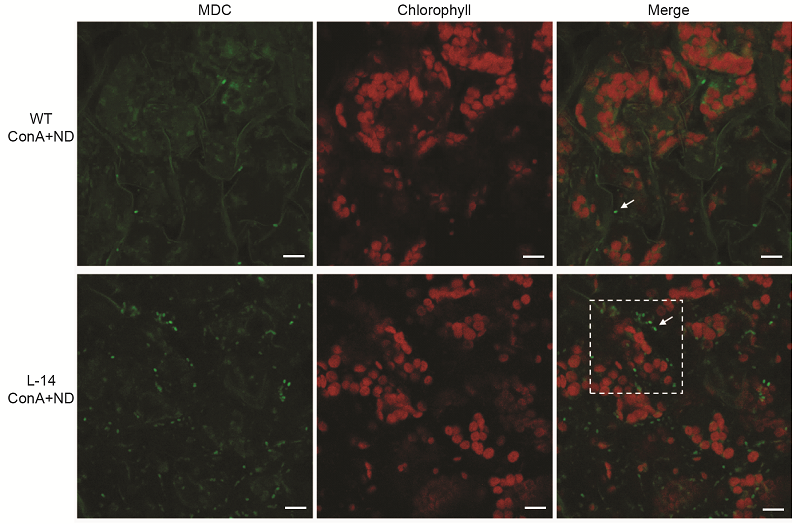
Figure 5 Overexpression of OsATG8b in Arabidopsis enhanced the autophagic activity under N deficient condition 7-day-old seedlings of transgenic line (L-14) and wild type (WT) were transferred to in N-deficient (ND) liquid medium with 1 μmol·L-1 ConA for 12 h, MDC-stained autophagosomes in leaves were observed by confocal microscopy. Bars=10 μm
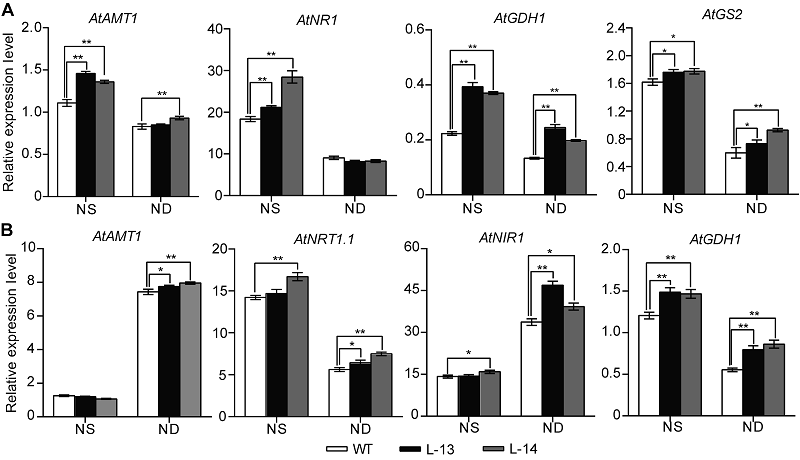
Figure 6 Overexpression of OsATG8b in Arabidopsis changes the expression of genes in nitrogen metabolic 7-day-old seedlings of transgenic lines and wild type (WT) were transferred to 1/2MS medium with sufficient (NS) or deficient (ND) nitrogen for 14 days. (A) The expression of genes related to nitrogen metabolic in rosette leaves of 35S-OsATG8b transgenic lines and WT; (B) The expression of genes related to nitrogen metabolic in roots of 35S-OsATG8b transgenic lines and WT. Values are means±SD, n=10, * and ** indicate significant (P<0.05) and extremely significant (P<0.01) differences between transgenic lines and WT (Student’s t-test), respectively, three biological replicates were performed.
| 1 |
黄晓, 李发强 ( 2016). 细胞自噬在植物细胞程序性死亡中的作用. 植物学报 51, 859-862.
DOI URL |
| 2 |
景红娟, 周广舟, 谭晓荣, 平康康, 任雪建 ( 2012). 活性氧对植物自噬调控的研究进展. 植物学报 47, 534-542.
DOI URL |
| 3 | 刘洋, 张静, 王秋玲, 侯岁稳 ( 2018). 植物细胞自噬研究进展. 植物学报 53, 5-16. |
| 4 | 任晨霞, 龚清秋 ( 2014). 细胞自噬在植物碳氮营养中作用的研究进展. 中国细胞生物学学报 36, 407-414. |
| 5 | Arnon DI ( 1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris.Plant Physiol 24, 1-15. |
| 6 | Avila-Ospina L, Moison M, Yoshimoto K, Masclaux- Daubresse C ( 2014). Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65, 3799-3811. |
| 7 |
Biederbick A, Kern HF, Elsässer HP ( 1995). Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles.Eur J Cell Biol 66, 3-14.
DOI URL PMID |
| 8 |
Bradford MM ( 1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248-254.
DOI URL PMID |
| 9 | Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, Zhang CJ, Morris K, Jenner C, Jackson S, Thomas B, Tabrett A, Legaie R, Moore JD, Wild DL, Ott S, Rand D, Beynon J, Denby K, Mead A, Buchanan-Wollaston V ( 2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873-894. |
| 10 |
Chardon F, Noël V, Masclaux-Daubresse C ( 2012). Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J Exp Bot 63, 3401-3412.
DOI URL PMID |
| 11 | Clough SJ, Bent AF ( 1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana.Plant J 16, 735-743. |
| 12 |
Contento AL, Xiong Y, Bassham DC ( 2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J 42, 598-608.
DOI URL PMID |
| 13 | Feng YC, He D, Yao ZY, Klionsky DJ ( 2014). The machi- nery of macroautophagy. Cell Res 24, 24-41. |
| 14 |
Good AG, Shrawat AK, Muench DG ( 2004). Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9, 597-605.
DOI URL PMID |
| 15 | Guiboileau A, Avila-Ospina L, Yoshimoto K, Soulay F, Azzopardi M, Marmagne A, Lothier J, Masclaux- Daubresse C ( 2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol 199, 683-694. |
| 16 | Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C ( 2012). Autophagy machi- nery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194, 732-740. |
| 17 |
Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T ( 2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process.Plant Physiol 148, 142-155.
DOI URL |
| 18 |
Izumi M, Hidema J, Makino A, Ishida H ( 2013). Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol 161, 1682-1693.
DOI URL PMID |
| 19 |
Izumi M, Hidema J, Wada S, Kondo E, Kurusu T, Kuchitsu K, Makino A, Ishida H ( 2015). Establishment of monito- ring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol 167, 1307-1320.
DOI URL PMID |
| 20 |
Kichey T, Hirel B, Heumez E, Dubois F, Le Gouis J ( 2007). In winter wheat ( Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers.Field Crops Res 102, 22-32.
DOI URL |
| 21 | Kraiser T, Gras DE, Gutiérrez AG, González B, Gutiérrez RA ( 2011). A holistic view of nitrogen acquisition in plants. J Exp Bot 62, 1455-1466. |
| 22 |
Krapp A ( 2015). Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Curr Opin Plant Biol 25, 115-122.
DOI URL PMID |
| 23 |
Li FQ, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD ( 2015 a). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389-1408.
DOI URL PMID |
| 24 |
Li WW, Chen M, Wang EH, Hu LQ, Hawkesford MJ, Zhong L, Chen Z, Xu ZS, Li LC, Zhou YB, Guo CH, Ma YZ ( 2016). Genome-wide analysis of autophagy-associated genes in foxtail millet ( Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice.BMC Genomics 17, 797.
DOI URL PMID |
| 25 | Li WW, Chen M, Zhong L, Liu JM, Xu ZS, Li LC, Zhou YB, Guo CH, Ma YZ ( 2015 b). Overexpression of the autophagy-related gene SiATG8a from foxtail millet( Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem Biophys Res Com- mun 468, 800-806. |
| 26 |
Liu D, Gong QQ, Ma YY, Li PL, Li JP, Yang SH, Yuan LL, Yu YQ, Pan DD, Xu F, Wang NN ( 2010). Cpseca, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J Exp Bot 61, 1655-1669.
DOI URL PMID |
| 27 |
Liu YM, Bassham DC ( 2012). Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63, 215-237.
DOI URL PMID |
| 28 |
Makino A, Osmond B ( 1991). Effects of nitrogen nutrition on nitrogen partitioning between chloroplasts and mitochondria in pea and wheat. Plant Physiol 96, 355-362.
DOI URL PMID |
| 29 |
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A ( 2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105, 1141-1157.
DOI URL PMID |
| 30 |
Masclaux-Daubresse C, Reisdorf-Cren M, Orsel M ( 2008). Leaf nitrogen remobilisation for plant development and gr- ain filling. Plant Biol 10, 23-36.
DOI URL PMID |
| 31 | Meyer C, Stitt M ( 2001). Nitrate reduction and signaling. In: Lea PJ, Morot-Gaudry JF, eds. Plant Nitrogen. Berlin, Heidelberg: Springer. pp. 37-59. |
| 32 |
Moriyasu Y, Ohsumi Y ( 1996). Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol 111, 1233-1241.
DOI URL PMID |
| 33 | Ohsumi Y ( 2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2, 211-216. |
| 34 |
Otegui MS, Noh YS, Martínez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ ( 2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41, 831-844.
DOI URL PMID |
| 35 |
Patrick JW, Offler CE ( 2001). Compartmentation of transport and transfer events in developing seeds. J Exp Bot 52, 551-564.
DOI URL PMID |
| 36 |
Rentsch D, Schmidt S, Tegeder M ( 2007). Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 581, 2281-2289.
DOI URL PMID |
| 37 |
Roberts IN, Caputo C, Criado MV, Funk C ( 2012). Senescence-associated proteases in plants. Physiol Plant 145, 130-139.
DOI URL PMID |
| 38 |
Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G ( 2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J Exp Bot 59, 4029-4043.
DOI URL PMID |
| 39 | Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD ( 2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways.Plant Physiol 138, 2097-2110. |
| 40 | Tsukada M, Ohsumi Y ( 1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae.FEBS Lett 333, 169-174. |
| 41 | Wada S, Hayashida Y, Izumi M, Kurusu T, Hanamata S, Kanno K, Kojima S, Yamaya T, Kuchitsu K, Makino A, Ishida H ( 2015). Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol 168, 60-73. |
| 42 |
Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A ( 2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149, 885-893.
DOI URL |
| 43 |
Walch-Liu P, Filleur S, Gan YB, Forde BG ( 2005). Signaling mechanisms integrating root and shoot responses to ch- anges in the nitrogen supply. Photosynth Res 83, 239-250.
DOI URL PMID |
| 44 |
Wang P, Sun X, Jia X, Wang N, Gong XQ, Ma FW ( 2016). Characterization of an autophagy-related gene MdATG8i from apple.Front Plant Sci 7, 720.
DOI URL PMID |
| 45 | Wang Y, Yu BJ, Zhao JP, Guo JB, Li Y, Han SJ, Huang L, Du YM, Hong YG, Tang DZ, Liu YL ( 2013). Autophagy contributes to leaf starch degradation. Plant Cell 25, 1383-1399. |
| 46 |
Xia KF, Liu T, Ouyang J, Wang R, Fan T, Zhang MY ( 2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice ( Oryza sativa L.).DNA Res 18, 363-377.
DOI URL PMID |
| 47 | Xia TM, Xiao D, Liu D, Chai WT, Gong QQ, Wang NN ( 2012). Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis.PLoS One 7, e37217. |
| 48 |
Yang XC, Bassham DC ( 2015). New insight into the mechanism and function of autophagy in plant cells. Int Rev Cell Mol Biol 320, 1-40.
DOI URL PMID |
| 49 |
Yao ZY, Delorme-Axford E, Backues SK, Klionsky DJ ( 2015). Atg41/Icy2 regulates autophagosome formation. Autophagy 11, 2288-2299.
DOI URL PMID |
| 50 |
Yoshimoto K ( 2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53, 1355-1365.
DOI URL PMID |
| 51 |
Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y ( 2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967-2983.
DOI URL PMID |
| 52 | Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, OhsumiY, Shirasu K ( 2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914-2927. |
| [1] | TANG Yuan-Xiang, XIONG Shi-Chen, ZHU Hong-Feng, ZHANG Xin-Sheng, YOU Cheng-Ming, LIU Si-Ning, TAN Bo, XU Zhen-Feng. Effects of long-term nitrogen addition on leaf litter production and carbon, nitrogen and phosphorus return of the dominant tree species in broadleaf evergreen forests on the western margin of Sichuan Basin [J]. Chin J Plant Ecol, 2025, 49(5): 720-731. |
| [2] | Xu Tingyang, Liu Yuchen, Wang Wanpeng, Su Hang, Su Kunlong, Wu Zhenying, Lϋ Ming, Li Fuli, Wang Xiaoshan, Fu Chunxiang. Effects of Different Plant Growth Regulators on Wheat Growth and Development in the Saline-alkali Land [J]. Chinese Bulletin of Botany, 2025, 60(3): 354-362. |
| [3] | Chaoyu Zhu, Chengxiang Hu, Zhenan Zhu, Zhining Zhang, Lihai Wang, Jun Chen, Sanfeng Li, Jinjin Lian, Luyao Tang, Qianqian Zhong, Wenjing Yin, Yuexing Wang, Yuchun Rao. Mapping of QTLs Associated with Rice Panicle Traits and Candidate Gene Analysis [J]. Chinese Bulletin of Botany, 2024, 59(2): 217-230. |
| [4] | Yuping Yan, Xiaoqi Yu, Deyong Ren, Qian Qian. Genetic Mechanisms and Breeding Utilization of Grain Number Per Panicle in Rice [J]. Chinese Bulletin of Botany, 2023, 58(3): 359-372. |
| [5] | LIU Jian-Xin, LIU Rui-Rui, LIU Xiu-Li, JIA Hai-Yan, BU Ting, LI Na. Regulation of exogenous hydrogen sulfide on photosynthetic carbon metabolism in Avena nude under saline-alkaline stress [J]. Chin J Plant Ecol, 2023, 47(3): 374-388. |
| [6] | Yuqiang Liu, Jianmin Wan. The Host Controls the Protein Level of Insect Effectors to Balance Immunity and Growth [J]. Chinese Bulletin of Botany, 2023, 58(3): 353-355. |
| [7] | Weijun Ye, Yin Zhang, Peiran Wang, Lingling Zhang, Dongfeng Tian, Zejiang Wu, Bin Zhou. QTLs Analysis for Five Yield-related Traits in Mungbean [J]. Chinese Bulletin of Botany, 2023, 58(1): 150-158. |
| [8] | YU Shui-Jin, WANG Juan, ZHANG Chun-Yu, ZHAO Xiu-Hai. Impact and mechanism of maintaining biomass stability in a temperate coniferous and broadleaved mixed forest [J]. Chin J Plant Ecol, 2022, 46(6): 632-641. |
| [9] | Liu Xiaolong, Ji Ping, Yang Hongtao, Ding Yongdian, Fu Jialing, Liang Jiangxia, Yu Congcong. Priming Effect of Abscisic Acid on High Temperature Stress During Rice Heading-flowering Stage [J]. Chinese Bulletin of Botany, 2022, 57(5): 596-610. |
| [10] | Wang Lei, Chong Kang. Choice of both Ways: Variations of Reverted Repeats Balance Environmental Adaptation and Yield in Maize [J]. Chinese Bulletin of Botany, 2022, 57(5): 555-558. |
| [11] | XIONG Shu-Ping, CAO Wen-Bo, CAO Rui, ZHANG Zhi-Yong, FU Xin-Lu, XU Sai-Jun, PAN Hu-Qiang, WANG Xiao-Chun, MA Xin-Ming. Effects of horizontal structure on canopy vertical structure, microenvironment and yield of Triticum aestivum [J]. Chin J Plant Ecol, 2022, 46(2): 188-196. |
| [12] | LIN Yong, CHEN Zhi, YANG Meng, CHEN Shi-Ping, GAO Yan-Hong, LIU Ran, HAO Yan-Bin, XIN Xiao-Ping, ZHOU Li, YU Gui-Rui. Temporal and spatial variations of ecosystem photosynthetic parameters in arid and semi-arid areas of China and its influencing factors [J]. Chin J Plant Ecol, 2022, 46(12): 1461-1472. |
| [13] | Jian-Min Zhou. A Ca2+-ROS Signaling Axis in Rice Provides Clues to Rice-pathogen Coevolution and Crop Improvements [J]. Chinese Bulletin of Botany, 2021, 56(5): 513-515. |
| [14] | Xibao Li, Minyi Lai, Shan Liang, Xiaojing Wang, Caiji Gao, Chao Yang. Function and Transcriptional Regulation of Autophagy-related Genes in Plants [J]. Chinese Bulletin of Botany, 2021, 56(2): 201-217. |
| [15] | LI Zhou-Yuan, YE Xiao-Zhou, WANG Shao-Peng. Ecosystem stability and its relationship with biodiversity [J]. Chin J Plant Ecol, 2021, 45(10): 1127-1139. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||