

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (1): 94-103.DOI: 10.11983/CBB16247 cstr: 32102.14.CBB16247
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Qiaoli Li, Na Yan, Qiong Song, Junzhan Guo*( )
)
Received:2016-12-13
Accepted:2017-03-29
Online:2018-01-01
Published:2018-08-10
Contact:
Junzhan Guo
Qiaoli Li, Na Yan, Qiong Song, Junzhan Guo. Complete Chloroplast Genome Sequence and Characteristics Analysis of Morus multicaulis[J]. Chinese Bulletin of Botany, 2018, 53(1): 94-103.
| Genome feature | Morus indica | M. mongolica | M. notabilis | M. multicaulis |
|---|---|---|---|---|
| Genome size (bp) | 158484 | 158459 | 158680 | 159154 |
| LSC length (bp)/percent (%)/GC content (%) | 87386/55.14/34.1 | 87367/55.14/34.0 | 87470/55.12/34.1 | 87763/55.15/33.9 |
| SSC length (bp)/percent (%)/GC content (%) | 19742/12.46/29.4 | 19736/12.45/29.3 | 19776/12.46/29.3 | 20035/12.59/29.3 |
| IR length (bp)/percent (%)/GC content (%) | 25678/16.20/42.9 | 25678/16.20/42.9 | 25717/16.21/42.9 | 25678/16.13/42.9 |
| GC content (%) | 36.4 | 36.3 | 36.4 | 36.2 |
| Number of genes | 133 | 133 | 129 | 130 |
| Number of protein-coding genes | 88 | 88 | 84 | 85 |
Table 1 Comparison of chloroplast genomes among four species of Morus
| Genome feature | Morus indica | M. mongolica | M. notabilis | M. multicaulis |
|---|---|---|---|---|
| Genome size (bp) | 158484 | 158459 | 158680 | 159154 |
| LSC length (bp)/percent (%)/GC content (%) | 87386/55.14/34.1 | 87367/55.14/34.0 | 87470/55.12/34.1 | 87763/55.15/33.9 |
| SSC length (bp)/percent (%)/GC content (%) | 19742/12.46/29.4 | 19736/12.45/29.3 | 19776/12.46/29.3 | 20035/12.59/29.3 |
| IR length (bp)/percent (%)/GC content (%) | 25678/16.20/42.9 | 25678/16.20/42.9 | 25717/16.21/42.9 | 25678/16.13/42.9 |
| GC content (%) | 36.4 | 36.3 | 36.4 | 36.2 |
| Number of genes | 133 | 133 | 129 | 130 |
| Number of protein-coding genes | 88 | 88 | 84 | 85 |
| Function | Gene group | Gene name | |||
|---|---|---|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4 | rrn5 | rrn16 | rrn23 |
| Transfer RNA genes | trnA-UGC trnF-GAA trnH-GUG trnL-CAA trnN-GUU trnR-UCU trnT-GGU trnW-CCA | trnC-GCA trnfM-CAU trnI-CAU trnL-UAA trnP-UGG trnS-GCU trnT-UGU trnY-GUA | trnD-GUC trnG-GCC trnI-GAU trnL-UAG trnQ-UUG trnS-GGA trnV-GAC | trnE-UUC trnG-UCC trnK-UUU trnM-CAU trnR-ACG trnS-UGA trnV-UAC | |
| Small subunit of ribosome | rps2 rps8 rps15 | rps3 rps11 rps16* | rps4 rps12 rps18 | rps7 rps14 rps19 | |
| Lange subunit of ribosome | rpl2* rpl22 rpl36 | rpl14 rpl23 | rpl16* rpl32 | rpl20 rpl33 | |
| RNA polymerase subunits | rpoA | rpoB | rpoC1* | rpoC2 | |
| NADH dehydrogenase | ndhA* ndhE ndhI | ndhB* ndhF ndhJ | ndhC ndhG ndhK | ndhD ndhH | |
| Photosynthesis | Photosystem I | psaA psaJ | psaB | psaC | psaI |
| Photosystem II | psbA psbE psbJ psbN | psbB psbF psbK psbT | psbC psbH psbL psbZ | psbD psbI psbM | |
| Cytochrome b/f complex | petA petL | petB* petN | petD* | petG | |
| ATP synthase | atpA atpH | atpB atpI | atpE | atpF* | |
| ATP Protease | rbcl | ||||
| Large subunit of rubisco | matK | ||||
| Maturase | clpP* | ||||
| Envelope membrane protein | cemA | ||||
| Other genes | Subunit of acetyl-CoA-carboxylase | accD | |||
| C-type cytochrome synthesis | ccsA | ||||
| Unknown function | Hypothetical chloroplast reading frames | yf1 | ycf3* | ycf4 | ycf15 |
| ORFs | ycf2 ycf68* | ||||
Table 2 Genes present in the chloroplast genome of Morus multicaulis
| Function | Gene group | Gene name | |||
|---|---|---|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4 | rrn5 | rrn16 | rrn23 |
| Transfer RNA genes | trnA-UGC trnF-GAA trnH-GUG trnL-CAA trnN-GUU trnR-UCU trnT-GGU trnW-CCA | trnC-GCA trnfM-CAU trnI-CAU trnL-UAA trnP-UGG trnS-GCU trnT-UGU trnY-GUA | trnD-GUC trnG-GCC trnI-GAU trnL-UAG trnQ-UUG trnS-GGA trnV-GAC | trnE-UUC trnG-UCC trnK-UUU trnM-CAU trnR-ACG trnS-UGA trnV-UAC | |
| Small subunit of ribosome | rps2 rps8 rps15 | rps3 rps11 rps16* | rps4 rps12 rps18 | rps7 rps14 rps19 | |
| Lange subunit of ribosome | rpl2* rpl22 rpl36 | rpl14 rpl23 | rpl16* rpl32 | rpl20 rpl33 | |
| RNA polymerase subunits | rpoA | rpoB | rpoC1* | rpoC2 | |
| NADH dehydrogenase | ndhA* ndhE ndhI | ndhB* ndhF ndhJ | ndhC ndhG ndhK | ndhD ndhH | |
| Photosynthesis | Photosystem I | psaA psaJ | psaB | psaC | psaI |
| Photosystem II | psbA psbE psbJ psbN | psbB psbF psbK psbT | psbC psbH psbL psbZ | psbD psbI psbM | |
| Cytochrome b/f complex | petA petL | petB* petN | petD* | petG | |
| ATP synthase | atpA atpH | atpB atpI | atpE | atpF* | |
| ATP Protease | rbcl | ||||
| Large subunit of rubisco | matK | ||||
| Maturase | clpP* | ||||
| Envelope membrane protein | cemA | ||||
| Other genes | Subunit of acetyl-CoA-carboxylase | accD | |||
| C-type cytochrome synthesis | ccsA | ||||
| Unknown function | Hypothetical chloroplast reading frames | yf1 | ycf3* | ycf4 | ycf15 |
| ORFs | ycf2 ycf68* | ||||
| Codon | Amino acid | Number | Codon | Amino acid | Number |
|---|---|---|---|---|---|
| GGG | Gly(G) | 494 | TGG | Trp(W) | 684 |
| GGA | Gly(G) | 759 | TGA | Stop | 1032 |
| GGT | Gly(G) | 599 | TGT | Cys(C) | 725 |
| GGC | Gly(G) | 350 | TGC | Cys(C) | 435 |
| GAG | Glu(E) | 550 | TAG | Stop | 786 |
| GAA | Glu(E) | 1368 | TAA | Stop | 1306 |
| GAT | Asp(D) | 1064 | TAT | Try(Y) | 1624 |
| GAC | Asp(D) | 425 | TAC | Try(Y) | 690 |
| GTG | Val(V) | 418 | TTG | Leu(L) | 1073 |
| GTA | Val(V) | 728 | TTA | Leu(L) | 1250 |
| GTT | Val(V) | 792 | TTT | Phe(F) | 2343 |
| GTC | Val(V) | 430 | TTC | Phe(F) | 1471 |
| GCG | Ala(A) | 249 | TCG | Ser(S) | 578 |
| GCA | Ala(A) | 430 | TCA | Ser(S) | 979 |
| GCT | Ala(A) | 511 | TCT | Ser(S) | 1273 |
| GCC | Ala(A) | 321 | TCC | Ser(S) | 864 |
| AGG | Arg(R) | 596 | CGG | Arg(R) | 350 |
| AGA | Arg(R) | 1044 | CGA | Arg(R) | 596 |
| AGT | Ser(S) | 718 | CGT | Arg(R) | 363 |
| AGC | Ser(S) | 478 | CGC | Arg(R) | 236 |
| AAG | Lys(K) | 1039 | CAG | Gln(Q) | 440 |
| AAA | Lys(K) | 2280 | CAA | Gln(Q) | 1013 |
| AAT | Asn(N) | 1883 | CAT | His(H) | 945 |
| AAC | Asn(N) | 728 | CAC | His(H) | 362 |
| ATG | Met(M) | 855 | CTG | Leu(L) | 489 |
| ATA | Ile(I) | 1729 | CTA | Leu(L) | 799 |
| ATT | Ile(I) | 1965 | CTT | Leu(L) | 1065 |
| ATC | Ile(I) | 1083 | CTC | Leu(L) | 581 |
| ACG | Thr(T) | 399 | CCG | Pro(P) | 400 |
| ACA | Thr(T) | 689 | CCA | Pro(P) | 738 |
| ACT | Thr(T) | 690 | CCT | Pro(P) | 730 |
| ACC | Thr(T) | 587 | CCC | Pro(P) | 580 |
Table 3 Codon usage in Morus multicaulis
| Codon | Amino acid | Number | Codon | Amino acid | Number |
|---|---|---|---|---|---|
| GGG | Gly(G) | 494 | TGG | Trp(W) | 684 |
| GGA | Gly(G) | 759 | TGA | Stop | 1032 |
| GGT | Gly(G) | 599 | TGT | Cys(C) | 725 |
| GGC | Gly(G) | 350 | TGC | Cys(C) | 435 |
| GAG | Glu(E) | 550 | TAG | Stop | 786 |
| GAA | Glu(E) | 1368 | TAA | Stop | 1306 |
| GAT | Asp(D) | 1064 | TAT | Try(Y) | 1624 |
| GAC | Asp(D) | 425 | TAC | Try(Y) | 690 |
| GTG | Val(V) | 418 | TTG | Leu(L) | 1073 |
| GTA | Val(V) | 728 | TTA | Leu(L) | 1250 |
| GTT | Val(V) | 792 | TTT | Phe(F) | 2343 |
| GTC | Val(V) | 430 | TTC | Phe(F) | 1471 |
| GCG | Ala(A) | 249 | TCG | Ser(S) | 578 |
| GCA | Ala(A) | 430 | TCA | Ser(S) | 979 |
| GCT | Ala(A) | 511 | TCT | Ser(S) | 1273 |
| GCC | Ala(A) | 321 | TCC | Ser(S) | 864 |
| AGG | Arg(R) | 596 | CGG | Arg(R) | 350 |
| AGA | Arg(R) | 1044 | CGA | Arg(R) | 596 |
| AGT | Ser(S) | 718 | CGT | Arg(R) | 363 |
| AGC | Ser(S) | 478 | CGC | Arg(R) | 236 |
| AAG | Lys(K) | 1039 | CAG | Gln(Q) | 440 |
| AAA | Lys(K) | 2280 | CAA | Gln(Q) | 1013 |
| AAT | Asn(N) | 1883 | CAT | His(H) | 945 |
| AAC | Asn(N) | 728 | CAC | His(H) | 362 |
| ATG | Met(M) | 855 | CTG | Leu(L) | 489 |
| ATA | Ile(I) | 1729 | CTA | Leu(L) | 799 |
| ATT | Ile(I) | 1965 | CTT | Leu(L) | 1065 |
| ATC | Ile(I) | 1083 | CTC | Leu(L) | 581 |
| ACG | Thr(T) | 399 | CCG | Pro(P) | 400 |
| ACA | Thr(T) | 689 | CCA | Pro(P) | 738 |
| ACT | Thr(T) | 690 | CCT | Pro(P) | 730 |
| ACC | Thr(T) | 587 | CCC | Pro(P) | 580 |
| Length (bp) | Number | Morus multicaulis | M. mongolica |
|---|---|---|---|
| A10 | 10 | 2142, 3980, 5079, 5977, 29067, 49740, 68616, 68631, 114154 (ndhF), 116262 | 3760, 4859, 28847, 38118, 113758 (ndhF), 115866 |
| A11 | 3 | 9589, 62837, 87467 | 1921, 5757, 9371, 62504, 81011 |
| A12 | 3 | 4830, 53982, 85376 | 13368, 38142, 53676, 84178, 87070 |
| A13 | 1 | 13596 | 4609, 73766 |
| A14 | 1 | 128163 | 127468 |
| A15 | 1 | 74160 | |
| A16 | 1 | 8990 | |
| A17 | 8772 | ||
| T10 | 20 | 66, 5258, 8582, 9802, 14098, 14919, 24357, 30672, 30938, 54024, 57098 (atpB), 62610, 66927, 68743, 70892, 73958, 83130, 116784, 130487 (ycf1), 132244 (ycf1) | 5038, 7036, 9584, 24137, 30452, 30718, 53718, 56773 (atpB), 62277, 70506, 73564, 82753, 116369, 121665, 129792 (ycf1), 131549 (ycf1) |
| T11 | 6 | 513, 34264, 69552, 78684, 122351, 131346 (ycf1) | 293, 8363, 57218, 59233, 66594, 68126, 69166, 74280, 78285, 130651 (ycf1) |
| T12 | 5 | 27617 (rpoB), 57549, 59565, 72471, 85809 | 12476, 13966, 27397 (rpoB), 34035, 85411 |
| T13 | 5 | 12703, 13286, 68491, 81352, 128585 | 8996, 13058, 51524, 72085, 127890 |
| T14 | 5 | 9213, 51829, 63865, 74676, 86927 | 63532, 80953 |
| T16 | 49162, 86528 | ||
| T17 | 1 | 49475 | |
| T19 | 1 | 116631 | 116235 |
| AT5 | 1 | 11566 (ndhF) | 115270 (ndhF) |
| AT6 | 2 | 118643, 118871 | 10589, 49643 |
| TA6 | 2 | 5522, 21234 (rpoC2) | 5302, 21009 (rpoC2), 118243 |
| TC5 | 1 | 645927 (cemA) | 64259 (cemA) |
| TTC4 | 1 | 70909 | 70523 |
| AAT4 | 1 | 128565 | 127870 |
| ATTT3 | 1 | 62140 | |
| ATTT4 | 1 | 14187 | 13957, 61807 |
| AAAT3 | 2 | 24056 (rpoC1), 46731 (ycf3) | 23831(rpoC1), 46414 (ycf3) |
| TATT3 | 1 | 24388 (rpoC1) | 24168 (rpoC1) |
| ATTA3 | 2 | 33980, 116443 | 33751, 116047 |
| TCTT3 | 1 | 111575 | 111179 |
| AAAG3 | 1 | 135331 | 134636 |
| AAGGA3 | 1 | 14021 (atpF) | 13792 (atpF) |
| ATTTC3 | 24071 |
Table 4 Comparison of simple sequence repeats (SSR) loci in Morus multicaulis and M. mongolica
| Length (bp) | Number | Morus multicaulis | M. mongolica |
|---|---|---|---|
| A10 | 10 | 2142, 3980, 5079, 5977, 29067, 49740, 68616, 68631, 114154 (ndhF), 116262 | 3760, 4859, 28847, 38118, 113758 (ndhF), 115866 |
| A11 | 3 | 9589, 62837, 87467 | 1921, 5757, 9371, 62504, 81011 |
| A12 | 3 | 4830, 53982, 85376 | 13368, 38142, 53676, 84178, 87070 |
| A13 | 1 | 13596 | 4609, 73766 |
| A14 | 1 | 128163 | 127468 |
| A15 | 1 | 74160 | |
| A16 | 1 | 8990 | |
| A17 | 8772 | ||
| T10 | 20 | 66, 5258, 8582, 9802, 14098, 14919, 24357, 30672, 30938, 54024, 57098 (atpB), 62610, 66927, 68743, 70892, 73958, 83130, 116784, 130487 (ycf1), 132244 (ycf1) | 5038, 7036, 9584, 24137, 30452, 30718, 53718, 56773 (atpB), 62277, 70506, 73564, 82753, 116369, 121665, 129792 (ycf1), 131549 (ycf1) |
| T11 | 6 | 513, 34264, 69552, 78684, 122351, 131346 (ycf1) | 293, 8363, 57218, 59233, 66594, 68126, 69166, 74280, 78285, 130651 (ycf1) |
| T12 | 5 | 27617 (rpoB), 57549, 59565, 72471, 85809 | 12476, 13966, 27397 (rpoB), 34035, 85411 |
| T13 | 5 | 12703, 13286, 68491, 81352, 128585 | 8996, 13058, 51524, 72085, 127890 |
| T14 | 5 | 9213, 51829, 63865, 74676, 86927 | 63532, 80953 |
| T16 | 49162, 86528 | ||
| T17 | 1 | 49475 | |
| T19 | 1 | 116631 | 116235 |
| AT5 | 1 | 11566 (ndhF) | 115270 (ndhF) |
| AT6 | 2 | 118643, 118871 | 10589, 49643 |
| TA6 | 2 | 5522, 21234 (rpoC2) | 5302, 21009 (rpoC2), 118243 |
| TC5 | 1 | 645927 (cemA) | 64259 (cemA) |
| TTC4 | 1 | 70909 | 70523 |
| AAT4 | 1 | 128565 | 127870 |
| ATTT3 | 1 | 62140 | |
| ATTT4 | 1 | 14187 | 13957, 61807 |
| AAAT3 | 2 | 24056 (rpoC1), 46731 (ycf3) | 23831(rpoC1), 46414 (ycf3) |
| TATT3 | 1 | 24388 (rpoC1) | 24168 (rpoC1) |
| ATTA3 | 2 | 33980, 116443 | 33751, 116047 |
| TCTT3 | 1 | 111575 | 111179 |
| AAAG3 | 1 | 135331 | 134636 |
| AAGGA3 | 1 | 14021 (atpF) | 13792 (atpF) |
| ATTTC3 | 24071 |
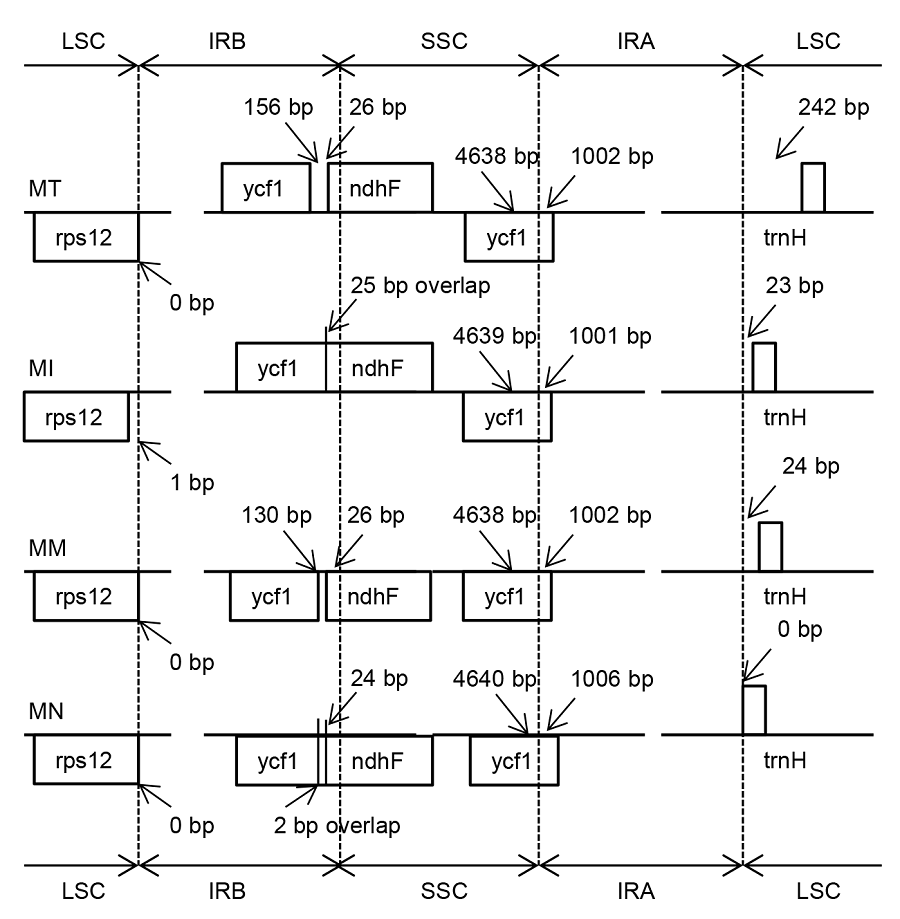
Figure 2 Comparison of the junction between inverted repeat region (IR), large single copy-region (LSC) and small single copy- region (SSC) of chloroplast genome among four Morus species MT: Morus multicaulis; MI: M. indica; MM: M. mongolica; MN: M. notabilis
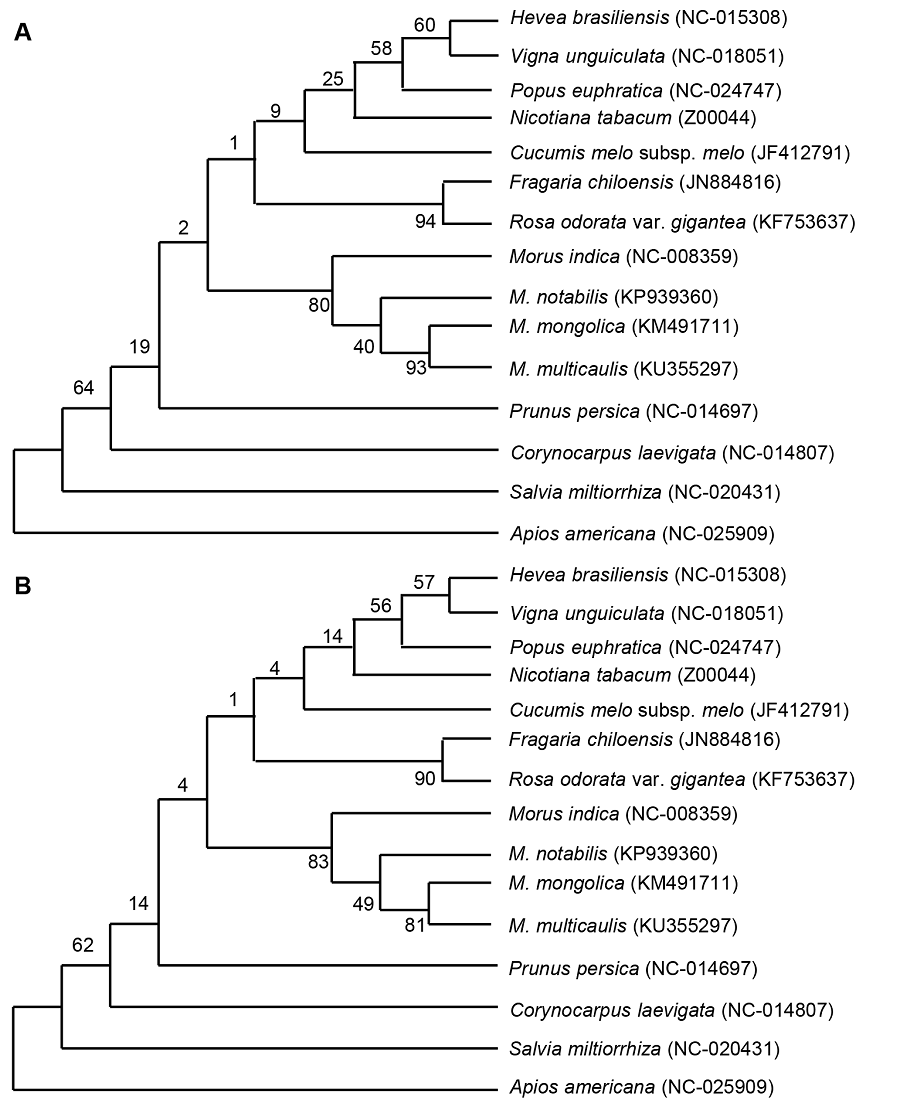
Figure 3 Cluster analysis of four species of Morus using complete chloroplast genome sequence by the maximum likelihood (ML) method (A) and neighbor-joining (NL) method (B)
| [1] | 冯丽春, 杨光伟, 余茂德, 张孝勇, 向怀祥 (1997). 利用RAPD对桑属植物种间亲缘关系的研究. 中国农业科学 30, 52-56. |
| [2] | 黄瑶, 李朝銮, 马诚, 吴乃虎 (1994). 叶绿体DNA及其在植物系统学研究中的应用. 植物学通报 11(2), 11-25. |
| [3] | 徐军望, 冯德江, 宋贵生, 魏晓丽, 陈蕾, 伍晓丽, 李旭刚, 朱桢 (2003). 水稻EPSP合酶第一内含子增强外源基因的表达. 中国科学(C辑) 33, 224-230. |
| [4] | 闫化学, 于杰 (2010). DNA条形码技术在植物中的研究现状. 植物学报 45, 102-108. |
| [5] | 周德贵, 赵琼一, 付崇允, 李宏, 蔡学飞, 罗达, 周少川 (2008). 新一代测序技术及其对水稻分子设计育种的影响. 分子植物育种 6, 619-630. |
| [6] | Allender CJ, Allainguillaume J, Lynn J, King GJ (2007). Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica ole- racea L. and(n=9) wild relatives. Theor Appl Genet 114, 609-618. |
| [7] | Chen C, Zhou W, Huang Y, Wang ZZ (2015). The complete chloroplast genome sequence of the mulberry Morus notabilis(Moreae). Mitochondrial DNA Part A 27, 2856-2857. |
| [8] | Flannery ML, Mitchell FJG, Coyne S, Kavanagh TA, Burke JI, Salamin N, Dowding P, Hodkinson TR (2006). Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor Appl Genet 113, 1221-1231. |
| [9] | George B, Bhatt BS, Awasthi M, George B, Singh AK (2015). Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants.Curr Genet 61, 665-677. |
| [10] | Hebert PD, Ratnasingham S, de Waard JR (2003). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B 270, S96-S99. |
| [11] | Huang YY, Matzke AJM, Matzke M (2013). Complete sequence and comparative analysis of the chloroplast genome of coconut palm ( Cocos nucifera). PLoS One 8, e74736. |
| [12] | Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ, Fourcade HM, Kuehl JV, McNeal JR, Leebens-Mack J, Cui LY (2005). Methods for obtaining and analyzing whole chloroplast genome sequen- ces.Methods Enzymol 395, 348-384. |
| [13] | Jiao Y, Jia HM, Li XW, Jia HJ, Chen Z, Wang GY, Chai CY, van de Weg E, Gao ZS (2012). Development of simple sequence repeat (SSR) markers from a genome survey of Chinese Bayberry ( Myrica rubra). BMC Genomics 13, 201. |
| [14] | Katti MV, Ranjekar PK, Gupta VS (2001). Differential distribution of simple sequence repeats in eukaryotic genome sequences.Mol Biol Evol 18, 1161-1167. |
| [15] | Kaundun SS, Matsumoto S (2002). Heterologous nuclear and chloroplast microsatellite amplification and variation in tea, Camellia sinensis. Genome 45, 1041-1048. |
| [16] | Kong WQ, Yang JH (2015). The complete chloroplast genome sequence of Morus mongolica and a comparative analysis within the Fabidae clade. Curr Genet 62, 165-172. |
| [17] | Leigh FJ, Mackay I, Oliveira HR, Gosman NE, Horsnell RA, Jones H, White J, Powell W, Brown TA (2013). Using diversity of the chloroplast genome to examine evolutionary history of wheat species.Genet Resour Crop Evol 60, 1831-1842. |
| [18] | Leseberg CH, Duvall MR (2009). The complete chloroplast genome of Coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals. J Mol Evol 69, 311-318. |
| [19] | Nazareno AG, Carlsen M, Lohmann LG (2015). Complete chloroplast genome of Tanaecium tetragonolobum: the first Bignoniaceae plastome. PLoS One 10, e0129930. |
| [20] | Plunkett GM, Downie SR (2000). Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae.Syst Bot 25, 648-667. |
| [21] | Rajendrakumar P, Biswal AK, Balachandran SM, Srinivasarao K, Sundaram RM (2007). Simple sequence repeats in organellar genomes of rice: frequency and distribution in genic and intergenic regions.Bioinformatics 23, 1-4. |
| [22] | Ravi V, Khurana JP, Tyagi AK, Khurana P (2006). The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis.Tree Genet Genomes 3, 49-59. |
| [23] | Ruhlman TA, Jansen RK (2014). The plastid genomes of flowering plants.Methods Mol Biol 1132, 3-38. |
| [24] | Shaw J, Lickey EB, Schilling EE, Small RL (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare lll.Am J Bot 94, 275-288. |
| [25] | Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001). Computational and experimental analysis of microsatellites in rice ( Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11, 1441-1452. |
| [26] | Zhang HY, Li C, Miao HM, Xiong SJ (2013). Insights from the complete chloroplast genome into the evolution of Se- samum indicum L. PLoS One 8, e80508. |
| [1] | Chuanyong Wang, Dian Zhuang, Zhengda Song, Henghua Zhai, Naiwei Li, Fan Zhang. Structural and Comparative Analysis of the Complete Chloroplast Genome and Phylogenetic Inference of the Aronia melanocarpa [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] | Shang Huadan, Zhang Chuqing, Wang Mei, Pei Wenya, Li Guohong, Wang Hongbin. Species diversity and geographic distribution of poplar pests in China [J]. Biodiv Sci, 2025, 33(2): 24370-. |
| [3] | Feng Zhang, Richard Dormatey, Yindu Liu, Chengju Li, Yunjiao Wang, Chunli Zhang, Ying Zhang, Youfang Fan, Panfeng Yao, Zhenzhen Bi, Yuhui Liu, Jiangping Bai, Chao Sun. Screening and Evaluation of Phosphite-tolerant Potatoes [J]. Chinese Bulletin of Botany, 2024, 59(4): 544-557. |
| [4] | Xiangzhang Wu, Fumin Lei, Yiyi Shan, Jing Yu. Distribution pattern of bryophyte diversity and environmental impact factors in urban parks of Shanghai [J]. Biodiv Sci, 2024, 32(2): 23364-. |
| [5] | Zhengming Luo, Jinxian Liu, Bianhua Zhang, Yanying Zhou, Aihua Hao, Kai Yang, Baofeng Chai. Diversity characteristics and driving factors of soil protist communities in subalpine meadow at different degradation stages [J]. Biodiv Sci, 2023, 31(8): 23136-. |
| [6] | ZHANG Zhong-Fu, WANG Si-Hai, YANG Wei, CHEN Jian. Response of rhizosphere microbial community structure and functional characteristics to health status of Malania oleifera [J]. Chin J Plant Ecol, 2023, 47(7): 1020-1031. |
| [7] | Yinger Mao, Xiumei Zhou, Nan Wang, Xiuxiu Li, Yuke You, Shangbin Bai. Impact of Phyllostachys edulis expansion to Chinese fir forest on the soil bacterial community [J]. Biodiv Sci, 2023, 31(6): 22659-. |
| [8] | BAI Xue, LI Yu-Jing, JING Xiu-Qing, ZHAO Xiao-Dong, CHANG Sha-Sha, JING Tao-Yu, LIU Jin-Ru, ZHAO Peng-Yu. Response mechanisms of millet and its rhizosphere soil microbial communities to chromium stress [J]. Chin J Plant Ecol, 2023, 47(3): 418-433. |
| [9] | Zhenzhou Chu, Gulbar Yisilam, Zezhong Qu, Xinmin Tian. Comparative Analyses on the Chloroplast Genome of Three Sympatric Atraphaxis Species [J]. Chinese Bulletin of Botany, 2023, 58(3): 417-432. |
| [10] | Wen Zhao, Dandan Wang, Mumin Reyila, Kaichuan Huang, Shun Liu, Baokai Cui. Soil microbial community structure of Larix gmelinii forest in the Aershan area [J]. Biodiv Sci, 2023, 31(2): 22258-. |
| [11] | Jinbo Bao, Zhijie Ding, Haoyu Miao, Xueli Li, Shuxian Ren, Ruoyan Jiao, Hao Li, Qianqian Deng, Yingzi Li, Xinmin Tian. Analysis of Chloroplast Genomes of Aleurites moluccana [J]. Chinese Bulletin of Botany, 2023, 58(2): 248-260. |
| [12] | Fan Xia, Jing Yang, Jian Li, Yang Shi, Lixin Gai, Wenhua Huang, Jingwei Zhang, Nan Yang, Fuli Gao, Yingying Han, Weidong Bao. Gut bacterial composition of four leopard cat subpopulations in Beijing [J]. Biodiv Sci, 2022, 30(9): 22103-. |
| [13] | Yixin Sun, Yingbin Li, Yuhui Li, Bing Li, Xiaofang Du, Qi Li. Application of high-throughput sequencing technique in the study of nematode diversity [J]. Biodiv Sci, 2022, 30(12): 22266-. |
| [14] | Cheng Gao, Liang-Dong Guo. Progress on microbial species diversity, community assembly and functional traits [J]. Biodiv Sci, 2022, 30(10): 22429-. |
| [15] | ZHOU Kai-Ling, ZHAO Yu-Jin, BAI Yong-Fei. Study on forest plant diversity monitoring based on Sentinel-2A satellite data in northeast China [J]. Chin J Plant Ecol, 2022, 46(10): 1251-1267. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||