

植物学报 ›› 2021, Vol. 56 ›› Issue (3): 330-338.DOI: 10.11983/CBB21057 cstr: 32102.14.CBB21057
收稿日期:2020-04-02
接受日期:2021-05-07
出版日期:2021-05-01
发布日期:2021-05-07
通讯作者:
张翠
作者简介:*E-mail: cuizhang@ibcas.ac.cn基金资助:
Binbin Hu1,2, Zhihui Xue1, Cui Zhang1,2,*( )
)
Received:2020-04-02
Accepted:2021-05-07
Online:2021-05-01
Published:2021-05-07
Contact:
Cui Zhang
摘要: 小RNA是对植物生长发育十分重要的一类小分子核苷酸, 在多种生命过程以及胁迫响应中发挥重要调控作用。对小RNA的定位研究有助于揭示它们的功能, 而小RNA荧光原位杂交(sRNA-FISH)是一种通过荧光检测技术对生物体内小 RNA进行定性或半定量分析的技术, 目前该技术已经在动物体内被广泛应用, 而在植物体内的应用还比较少。该文详细介绍了基于超高分辨率显微镜的sRNA-FISH的具体操作流程以及注意事项, 该技术可用于探究植物组织内小RNA的表达与定位。
胡滨滨, 薛治慧, 张翠. 植物小RNA荧光原位杂交实验方法. 植物学报, 2021, 56(3): 330-338.
Binbin Hu, Zhihui Xue, Cui Zhang. Protocols for Small RNA FISH in Plants. Chinese Bulletin of Botany, 2021, 56(3): 330-338.
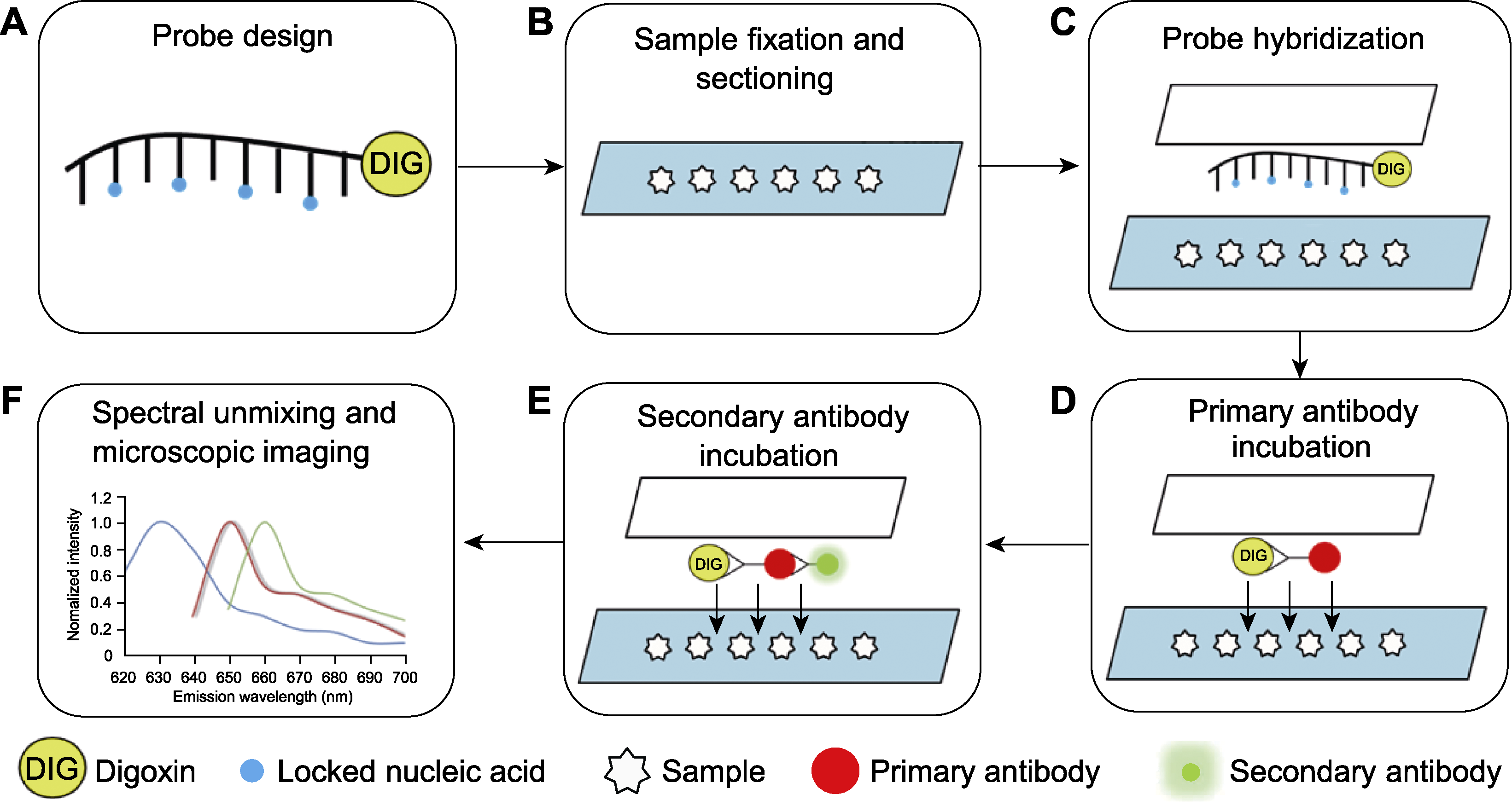
图1 小RNA荧光原位杂交实验流程图 (A) 探针设计; (B) 石蜡切片; (C) 探针杂交; (D) 一抗孵育; (E) 二抗孵育; (F) 光谱拆分以及激光共聚焦显微成像
Figure 1 Outlines of small RNA fluorescent in situ hybridization (A) Probe design; (B) Paraffin sectioning; (C) Probe hybridization; (D) Primary antibody incubation; (E) Secondary antibody incubation; (F) Spectral unmixing and imaging by confocal microscope
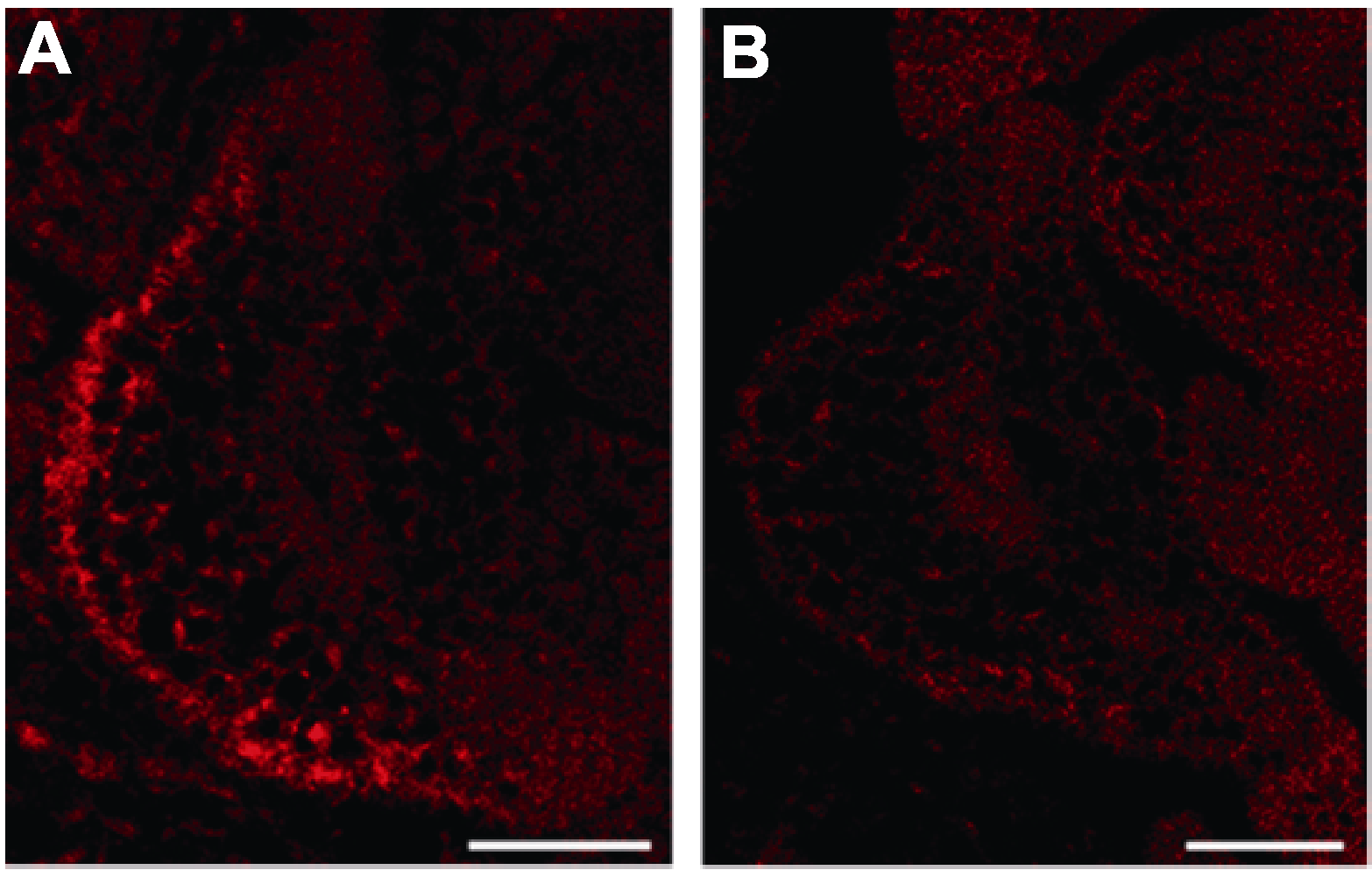
图2 小RNA FISH效果示例 (A) miR165/166反义寡核苷酸探针; (B) 阴性对照: miR165/ 166顺义寡核苷酸探针。Bars=50 μm
Figure 2 Schematic diagram of sRNA FISH (A) Antisense probe of miR165/166; (B) Negative control: Sense probe of miR165/166. Bars=50 μm
| 1 |
Baumberger N, Baulcombe DC (2005). Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102, 11928-11933.
DOI URL |
| 2 |
Bhogale S, Mahajan AS, Natarajan B, Rajabhoj M, Thulasiram HV, Banerjee AK (2014). MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol 164, 1011-1027.
DOI URL |
| 3 |
Bleckmann A, Dresselhaus T (2016). Fluorescent whole- mount RNA in situ hybridization (F-WISH) in plant germ cells and the fertilized ovule. Methods 98, 66-73.
DOI PMID |
| 4 |
Bruno L, Muto A, Spadafora ND, Iaria D, Chiappetta A, Van Lijsebettens M, Bitonti MB (2011). Multi-probe in situ hybridization to whole mount Arabidopsis seedlings. Int J Dev Biol 55, 197-203.
DOI URL |
| 5 |
Bruno L, Ronchini M, Gagliardi O, Corinti T, Chiappetta A, Gerola P, Bitonti MB (2015). Analysis of ATGUS1 and ATGUS2 in Arabidopsis root apex by a highly sensitive TSA-MISH method. Int J Dev Biol 59, 221-228.
DOI URL |
| 6 |
Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J (2008). Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J 53, 739-749.
DOI URL |
| 7 |
Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, Campilho A, Sebastian J, Bowman JL, Helariutta Y, Benfey PN (2010). Cell signaling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316-321.
DOI PMID |
| 8 |
Chen XM (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022-2025.
DOI URL |
| 9 |
Chen XM (2009). Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25, 21-44.
DOI URL |
| 10 | Chitwood DH, Nogueira FT, Howell MD, Montogmery TA, Carrington JC, Timmermans MC (2008). Pattern formation in leaves via small RNA mobility. Dev Biol 319, 587-588. |
| 11 |
Chitwood DH, Nogueira FTS, Howell MD, Montgomery TA, Carrington JC, Timmermans MCP (2009). Pattern formation via small RNA mobility. Gene Dev 23, 549-554.
DOI URL |
| 12 | Fischer AH, Jacobson KA, Rose J, Zeller R (2008). Paraffin embedding tissue samples for sectioning. CSH Protoc 2008, pdb.prot4989. |
| 13 |
Himanen K, Woloszynska M, Boccardi TM, De Groeve S, Nelissen H, Bruno L, Vuylsteke M, Van Lijsebettens M (2012). Histone H2b monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J 72, 249-260.
DOI URL |
| 14 |
Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidop- sis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19, 926-942.
PMID |
| 15 |
Huang K, Baldrich P, Meyers BC, Caplan JL (2019). sRNA-FISH: versatile fluorescent in situ detection of small RNAs in plants. Plant J 98, 359-369.
DOI |
| 16 |
Huen AK, Rodriguez-Medina C, Ho AYY, Atkins CA, Smith PMC (2017). Long-distance movement of phospha- te starvation-responsive microRNAs in Arabidopsis. Plant Biol 19, 643-649.
DOI URL |
| 17 |
Javelle M, Timmermans MCP (2012). In situ localization of small RNAs in plants by using LNA probes. Nat Protoc 7, 533-541.
DOI PMID |
| 18 | Jiang JM (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromoso- me Res 27, 153-165. |
| 19 |
Jones-Rhoades MW, Bartel DP, Bartel B (2006). Micro- RNAs and their regulatory roles in plants. Annu Rev Plant Biol 57, 19-53.
PMID |
| 20 |
Jung JH, Park CM (2007). MIR166/ 165 genes exhibit dynamic expression patterns in regulating shoot apical meris- tem and floral development in Arabidopsis. Planta 225, 1327-1338.
DOI URL |
| 21 | Kidner C, Timmermans M (2006). In situ hybridization as a tool to study the role of microRNAs in plant development. Methods Mol Biol 342, 159-179. |
| 22 | Kim VN, Han JJ, Siomi MC (2009). Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10, 126-139. |
| 23 |
Knauer S, Holt AL, Rubio-Somoza I, Tucker EJ, Hinze A, Pisch M, Javelle M, Timmermans MC, Tucker MR, Laux T (2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev Cell 24, 125-132.
DOI URL |
| 24 |
Kurihara Y, Watanabe Y (2004). Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc Natl Acad Sci USA 101, 12753-12758.
DOI URL |
| 25 |
Lamb JC, Danilova T, Bauer MJ, Meyer JM, Holland JJ, Jensen MD, Birchler JA (2007). Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175, 1047-1058.
DOI URL |
| 26 |
Lee Y, Kim M, Han JJ, Yeom KH, Lee S, Baek SH, Kim VN (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23, 4051-4060.
DOI URL |
| 27 |
Levsky JM, Singer RH (2003). Fluorescence in situ hybridization: past, present and future. J Cell Sci 116, 2833-2838.
DOI URL |
| 28 |
Li JJ, Yang ZY, Yu B, Liu J, Chen XM (2005). Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol 15, 1501-1507.
DOI URL |
| 29 |
Li S, Wang XT, Xu WY, Liu T, Cai CM, Chen LY, Clark CB, Ma JX (2021). Unidirectional movement of small RNAs from shoots to roots in interspecific heterografts. Nat Plants 7, 50-59.
DOI URL |
| 30 |
Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147, 732-746.
DOI URL |
| 31 |
Liu QL, Yao XZ, Pi LM, Wang H, Cui XF, Huang H (2009). The ARGONAUTE10 gene modulates shoot apical meris- tem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J 58, 27-40.
DOI URL |
| 32 |
Lu J, Tsourkas A (2009). Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res 37, e100.
DOI URL |
| 33 |
Moissiard G, Parizotto EA, Himber C, Voinnet O (2007). Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 13, 1268-1278.
DOI URL |
| 34 |
Nodine MD, Bartel DP (2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Gene Dev 24, 2678-2692.
DOI URL |
| 35 |
Nogueira FTS, Chitwood DH, Madi S, Ohtsu K, Schnable PS, Scanlon MJ, Timmermans MCP (2009). Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet 5, e1000320.
DOI URL |
| 36 |
Nogueira FTS, Madi S, Chitwood DH, Juarez MT, Timmer- mans MCP (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Gene Dev 21, 750-755.
PMID |
| 37 |
Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, Alvarez JP, Blum E, Zamir D, Eshed Y (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39, 787-791.
DOI URL |
| 38 |
Pagliarani C, Gambino G (2019). Small RNA mobility: spread of RNA silencing effectors and its effect on developmental processes and stress adaptation in plants. Int J Mol Sci 20, 4306.
DOI URL |
| 39 |
Pant BD, Buhtz A, Kehr J, Scheible WR (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53, 731-738.
DOI URL |
| 40 |
Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150, 1541-1555.
DOI URL |
| 41 |
Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Gene Dev 18, 2237-2242.
PMID |
| 42 |
Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA 102, 3691-3696.
DOI URL |
| 43 |
Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55, 65-76.
DOI URL |
| 44 |
Rozier F, Mirabet V, Vernoux T, Das P (2014). Analysis of 3D gene expression patterns in plants using whole-mount RNA in situ hybridization. Nat Protoc 9, 2464-2475.
DOI URL |
| 45 |
Skopelitis DS, Hill K, Klesen S, Marco CF, von Born P, Chitwood DH, Timmermans MCP (2018). Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat Commun 9, 3107.
DOI PMID |
| 46 |
Tirichine L, Andrey P, Biot E, Maurin Y, Gaudin V (2009). 3D fluorescent in situ hybridization using Arabidopsis leaf cryosections and isolated nuclei. Plant Methods 5, 11.
DOI PMID |
| 47 |
Vaucheret H, Vazquez F, Crété P, Bartel DP (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Gene Dev 18, 1187-1197.
PMID |
| 48 |
Wollmann H, Mica E, Todesco M, Long JA, Weigel D (2010). On reconciling the interactions between APETA- LA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633-3642.
DOI PMID |
| 49 |
Woloszynska M, Le Gall S, Neyt P, Boccardi TM, Grasser M, Längst G, Aesaert S, Coussens G, Dhondt S, Van De Slijke E, Bruno L, Fung-Uceda J, Mas P, Van Montagu M, Inzé D, Himanen K, De Jaeger G, Grasser KD, Van Lijsebettens M (2019). Histone 2B monoubiquitination complex integrates transcript elongation with RNA processing at circadian clock and flowering regulators. Proc Natl Acad Sci USA 116, 8060-8069.
DOI URL |
| 50 |
Xue ZH, Liu LY, Zhang C (2020). Regulation of shoot apical meristem and axillary meristem development in plants. Int J Mol Sci 21, 2917.
DOI URL |
| 51 |
Yang WB, Schuster C, Prunet N, Dong QK, Landrein B, Wightman R, Meyerowitz EM (2020). Visualization of protein coding, long noncoding, and nuclear RNAs by fluorescence in situ hybridization in sections of shoot apical meristems and developing flowers. Plant Physiol 182, 147-158.
DOI URL |
| 52 |
Yang WB, Wightman R, Meyerowitz EM (2017). Cell cycle control by nuclear sequestration of CDC20 and CDH1 mRNA in plant stem cells. Mol Cell 68, 1108-1119.
DOI URL |
| 53 |
Yu Y, Zhang YC, Chen XM, Chen YQ (2019). Plant noncoding RNAs: hidden players in development and stress responses. Annu Rev Cell Dev Biol 35, 407-431.
DOI |
| 54 |
Zhang C, Fan LS, Le BH, Ye PY, Mo BX, Chen XM (2020). Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis. Dev Cell 55, 603-616.
DOI URL |
| 55 | Zhang C, Wang J, Wenkel S, Chandler JW, Werr W, Jiao YL (2018). Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145, dev158352. |
| [1] | 陈龙 郭柯 勾晓华 赵秀海 马泓若. 祁连圆柏林群落组成及特征[J]. 植物生态学报, 2025, 49(植被): 0-0. |
| [2] | 惠城阳, 章巧依, 刘腾腾, 刘维勇, 周丽娜, 金鑫杰, 张永华, 刘金亮. 温州大罗山主要植被类型及物种组成特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [3] | 曹毅 张松林 王旭峰 杨安昌 任敏慧 杨浩 韩超. 兰州市南北两山植物群落数据集[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [4] | 童金莲, 张博纳, 汤璐瑶, 叶琳峰, 李姝雯, 谢江波, 李彦, 王忠媛. C4植物狗尾草功能性状网络沿降水梯度带的区域分异规律[J]. 植物生态学报, 2025, 49(预发表): 1-. |
| [5] | 闫小红 胡文海. 亚热带地区3种常绿阔叶植物冬季光保护机制的差异[J]. 植物生态学报, 2025, 49(预发表): 0-0. |
| [6] | 赵常明 熊高明 申国珍 葛结林 徐文婷 徐凯 武元帅 谢宗强. 神农架常绿落叶阔叶混交林和亚高山针叶林植物群落特征数据集[J]. 植物生态学报, 2025, 49(典型生态系统数据集): 0-0. |
| [7] | 赵珮杉 高广磊 丁国栋 张英. 林龄和生态位对樟子松人工林地下真菌群落构建的影响[J]. 植物生态学报, 2025, 49(地上地下生态过程关联): 1-0. |
| [8] | 黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群野生植物荔波杜鹃保护遗传学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [9] | 周鑫宇, 刘会良, 高贝, 卢妤婷, 陶玲庆, 文晓虎, 张岚, 张元明. 新疆特有濒危植物雪白睡莲繁殖生物学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [10] | 高雨轩, 苏艳军, 冯育才, 张军, 汪小全, 刘玲莉. 珍稀濒危孑遗植物银杉的研究与保护现状[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [11] | 张子睿, 周静, 胡艳萍, 梁爽, 马永鹏, 陈伟乐. 极度濒危植物巧家五针松的根内和根际真菌群落特征[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [12] | 逯子佳, 王天瑞, 郑斯斯, 孟宏虎, 曹建国, Gregor Kozlowski, 宋以刚. 孑遗植物湖北枫杨的环境适应性遗传变异与遗传脆弱性[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [13] | 平晓燕, 杜毅倩, 赖仕蓉, 孔梦桥, 余国杰. 植物应对食草动物采食的化学防御策略研究进展[J]. 植物生态学报, 2025, 49(5): 667-680. |
| [14] | 贾妍妍, 柳华清, 解欣然, 王博, 张维, 杨允菲. 珍稀濒危植物天山梣林龄结构及种群动态[J]. 植物生态学报, 2025, 49(5): 760-772. |
| [15] | 朱润铖, 蔡锡安, 黄娟. 植物防御相关挥发性有机物排放及对氮沉降的响应[J]. 植物生态学报, 2025, 49(5): 681-696. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||