

植物学报 ›› 2020, Vol. 55 ›› Issue (4): 430-441.DOI: 10.11983/CBB19204 cstr: 32102.14.CBB19204
收稿日期:2019-10-18
接受日期:2020-04-26
出版日期:2020-07-01
发布日期:2020-05-21
通讯作者:
丛佩华
基金资助:
Caixia Zhang,Gaopeng Yuan,Xiaolei Han,Wuxing Li,Peihua Cong( )
)
Received:2019-10-18
Accepted:2020-04-26
Online:2020-07-01
Published:2020-05-21
Contact:
Peihua Cong
摘要: 为鉴定不同抗性苹果(Malus domestica)品种响应轮纹病菌胁迫的抗性相关蛋白表达差异, 以抗病品种华月及易感品种金冠为试材, 采用高通量同位素标记定量(IBT)技术结合液相色谱-串联质谱(LC-MS)鉴定技术, 对病原菌处理前后抗、感病品种叶片的蛋白质组差异表达进行分析, 共鉴定出171个差异表达蛋白(DEPs)。GO富集及KEGG通路分析表明, 在细胞组分、分子功能和生物过程3类中共注释到686个GO条目, 其中52个DEPs注释于KEGG通路的18个显著差异途径(P<0.05)。亚细胞定位预测分析表明, 171个DEPs中有170个分别定位于8类细胞器。蛋白功能注释分析表明, 46个DEPs注释于7类抗性相关蛋白, 包括类甜蛋白、过氧化物酶、多酚氧化酶、过敏原蛋白、几丁质酶、内切葡聚糖酶以及主乳胶蛋白。此外, 还对抗性相关蛋白的表达特点及基因定量结果进行了分析。该研究结果可为进一步解析抗、感病苹果品种应答轮纹病菌胁迫的抗性机制提供参考。
张彩霞,袁高鹏,韩晓蕾,李武兴,丛佩华. 不同抗性苹果品种应答轮纹病菌胁迫的差异蛋白质组分析. 植物学报, 2020, 55(4): 430-441.
Caixia Zhang,Gaopeng Yuan,Xiaolei Han,Wuxing Li,Peihua Cong. Proteome Analysis of Different Resistant Apple Cultivars in Response to the Stress of Ring Rot Disease. Chinese Bulletin of Botany, 2020, 55(4): 430-441.
| Accession No. | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| gi|657977120 | TCACCTTAGCCATCTTCTTCGC | TGCTAACTCGAACCCTGTGG |
| gi|658027651 | CTCCACTGTGCCTATTGCGA | GAGCCGGGTTAGCGAACAA |
| gi|658061109 | TTGATGCCAGCCCTGCAAAT | GGGCTTGAAGCTCGTTGTTG |
| gi|661567324 | GATTGCACCCCAGGCAATCA | TTGTGCTTCACGTAGCCGTA |
| gi|658009573 | AGGCATTCCCTCAGGACTAC | TTCCGACTTCATCCACTGC |
| MdActin | TGACCGAATGAGCAAGGAAATTACT | TACTCAGCTTTGGCAATCCACATC |
表1 荧光定量PCR引物序列
Table 1 Primers used in fluorescent quantitative PCR
| Accession No. | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| gi|657977120 | TCACCTTAGCCATCTTCTTCGC | TGCTAACTCGAACCCTGTGG |
| gi|658027651 | CTCCACTGTGCCTATTGCGA | GAGCCGGGTTAGCGAACAA |
| gi|658061109 | TTGATGCCAGCCCTGCAAAT | GGGCTTGAAGCTCGTTGTTG |
| gi|661567324 | GATTGCACCCCAGGCAATCA | TTGTGCTTCACGTAGCCGTA |
| gi|658009573 | AGGCATTCCCTCAGGACTAC | TTCCGACTTCATCCACTGC |
| MdActin | TGACCGAATGAGCAAGGAAATTACT | TACTCAGCTTTGGCAATCCACATC |

图1 抗、感病苹果品种华月和金冠应答轮纹病菌侵染的发病特征比较 R: 抗病品种华月; S: 感病品种金冠。Bars=1 cm
Figure 1 Comparison of morphological characteristics of resistant and susceptible apple cultivars Huayue and Golden Delicious in response to ring rot disease R: Resistant cultivar Huayue; S: Susceptible cultivar Golden Delicious. Bars=1 cm
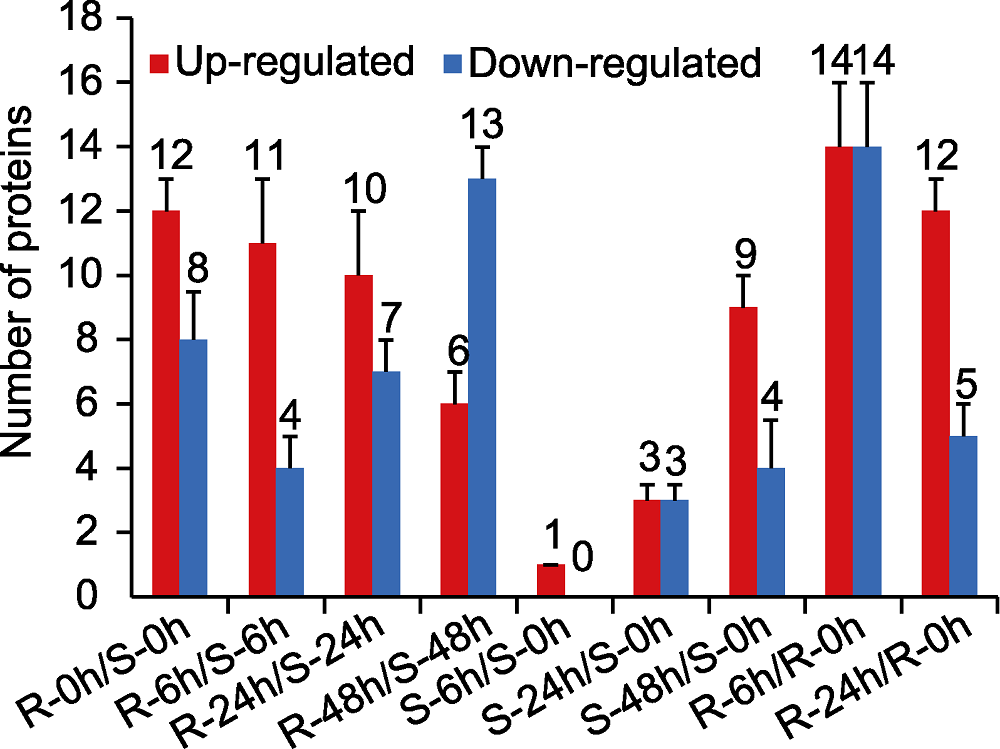
图2 抗、感病苹果品种叶片应答轮纹病菌胁迫的差异表达蛋白(DEPs)分布 R: 抗病品种华月; S: 感病品种金冠
Figure 2 Distribution of differentially expressed proteins (DEPs) of apple leaves susceptible and resistant to ring rot disease, respectively R: Resistant cultivar Huayue; S: Susceptible cultivar Golden Delicious

图3 抗、感病品种应答轮纹病菌胁迫的差异表达蛋白(DEPs)的Gene Ontology (GO)分析
Figure 3 Enriched Gene Ontology (GO) analysis of differentially expressed proteins (DEPs) in apple leaves resistant and susceptible to ring rot disease, respectively
| No. | Pathway | Pathway ID | DEPs with pathway annotation (326 in total) | All proteins with pathway annotation (14916 in total) | P-value |
|---|---|---|---|---|---|
| 1 | Fatty acid biosynthesis | ko00061 | 2 (0.61%) | 19 (0.13%) | 0.011 |
| 2 | Fatty acid elongation | ko00062 | 3 (0.92%) | 9 (0.06%) | 0.040 |
| 3 | Arginine biosynthesis | ko00220 | 2 (0.61%) | 16 (0.11%) | 0.050 |
| 4 | Alanine, aspartate and glutamate metabolism | ko00250 | 2 (0.61%) | 23 (0.15%) | 0.037 |
| 5 | Cyanoamino acid metabolism | ko00460 | 2 (0.61%) | 20 (0.13%) | 0.022 |
| 6 | Other glycan degradation | ko00511 | 2 (0.61%) | 14 (0.09%) | 0.011 |
| 7 | Monoterpenoid biosynthesis | ko00902 | 5 (1.53%) | 12 (0.08%) | 0.035 |
| 8 | Sesquiterpenoid and triterpenoid biosynthesis | ko00909 | 1 (0.31%) | 2 (0.01%) | 0.027 |
| 9 | Nitrogen metabolism | ko00910 | 4 (1.23%) | 28 (0.19%) | 0.014 |
| 10 | Sulfur metabolism | ko00920 | 1 (0.31%) | 12 (0.08%) | 0.036 |
| 11 | Phenylpropanoid biosynthesis | ko00940 | 3 (0.92%) | 47 (0.32%) | 0.041 |
| 12 | Isoquinoline alkaloid biosynthesis | ko00950 | 2 (0.61%) | 16 (0.11%) | 0.028 |
| 13 | Biosynthesis of unsaturated fatty acids | ko01040 | 2 (0.61%) | 9 (0.06%) | 0.016 |
| 14 | Fatty acid metabolism | ko01212 | 2 (0.61%) | 32 (0.21%) | 0.030 |
| 15 | Ribosome | ko03010 | 13 (3.99%) | 200 (1.34%) | 0.005 |
| 16 | RNA transport | ko03013 | 2 (0.61%) | 33 (0.22%) | 0.031 |
| 17 | Homologous recombination | ko03440 | 1 (0.31%) | 5 (0.03%) | 0.039 |
| 18 | Circadian rhythm-plant | ko04712 | 3 (0.92%) | 9 (0.06%) | 0.026 |
表2 差异表达蛋白(DEPs)的KEGG Pathway富集分析
Table 2 KEGG pathways analysis of differentially expressed proteins (DEPs)
| No. | Pathway | Pathway ID | DEPs with pathway annotation (326 in total) | All proteins with pathway annotation (14916 in total) | P-value |
|---|---|---|---|---|---|
| 1 | Fatty acid biosynthesis | ko00061 | 2 (0.61%) | 19 (0.13%) | 0.011 |
| 2 | Fatty acid elongation | ko00062 | 3 (0.92%) | 9 (0.06%) | 0.040 |
| 3 | Arginine biosynthesis | ko00220 | 2 (0.61%) | 16 (0.11%) | 0.050 |
| 4 | Alanine, aspartate and glutamate metabolism | ko00250 | 2 (0.61%) | 23 (0.15%) | 0.037 |
| 5 | Cyanoamino acid metabolism | ko00460 | 2 (0.61%) | 20 (0.13%) | 0.022 |
| 6 | Other glycan degradation | ko00511 | 2 (0.61%) | 14 (0.09%) | 0.011 |
| 7 | Monoterpenoid biosynthesis | ko00902 | 5 (1.53%) | 12 (0.08%) | 0.035 |
| 8 | Sesquiterpenoid and triterpenoid biosynthesis | ko00909 | 1 (0.31%) | 2 (0.01%) | 0.027 |
| 9 | Nitrogen metabolism | ko00910 | 4 (1.23%) | 28 (0.19%) | 0.014 |
| 10 | Sulfur metabolism | ko00920 | 1 (0.31%) | 12 (0.08%) | 0.036 |
| 11 | Phenylpropanoid biosynthesis | ko00940 | 3 (0.92%) | 47 (0.32%) | 0.041 |
| 12 | Isoquinoline alkaloid biosynthesis | ko00950 | 2 (0.61%) | 16 (0.11%) | 0.028 |
| 13 | Biosynthesis of unsaturated fatty acids | ko01040 | 2 (0.61%) | 9 (0.06%) | 0.016 |
| 14 | Fatty acid metabolism | ko01212 | 2 (0.61%) | 32 (0.21%) | 0.030 |
| 15 | Ribosome | ko03010 | 13 (3.99%) | 200 (1.34%) | 0.005 |
| 16 | RNA transport | ko03013 | 2 (0.61%) | 33 (0.22%) | 0.031 |
| 17 | Homologous recombination | ko03440 | 1 (0.31%) | 5 (0.03%) | 0.039 |
| 18 | Circadian rhythm-plant | ko04712 | 3 (0.92%) | 9 (0.06%) | 0.026 |
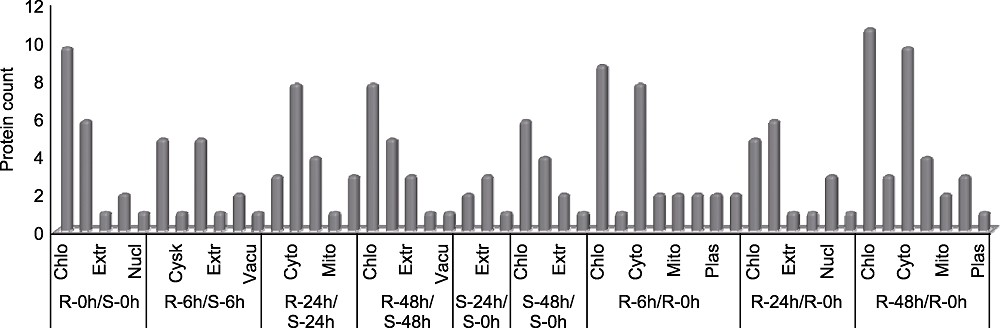
图4 抗、感病苹果叶片应答轮纹病菌胁迫的差异表达蛋白(DEPs)的亚细胞定位 Chlo: 叶绿体; Cysk: 细胞骨架; Cyto: 胞液; Extr: 胞外; Mito: 线粒体; Nucl: 细胞核; Plas: 质膜; Vacu: 液泡膜
Figure 4 Subcellular localization of differentially expressed proteins (DEPs) in apple leaves resistant and susceptible to ring rot disease, respectively Chlo: Chloroplast; Cysk: Cytoskeleton; Cyto: Cytosol; Extr: Extracellular; Mito: Mitochondria; Nucl: Nucleus; Plas: Plasma membrane; Vacu: Vacuolar membrane
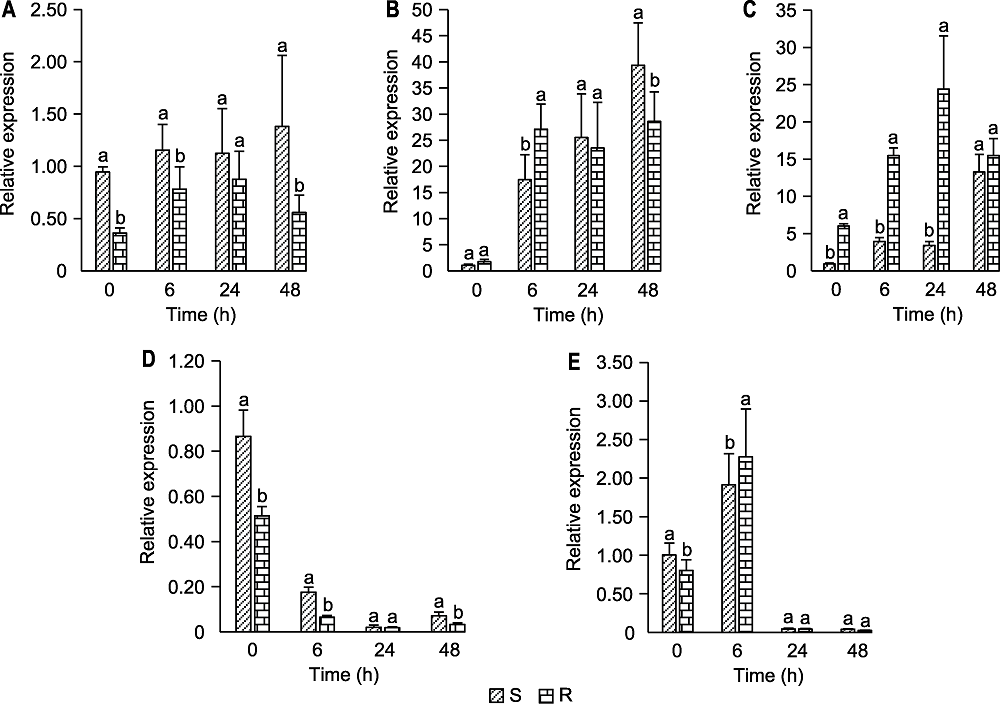
图5 抗、感病苹果叶片轮纹病关键抗病相关蛋白编码基因的实时荧光定量PCR分析 (A) 类甜蛋白编码基因(MD04G1018400); (B) Mal d1编码基因(MD16G1160700); (C) β-1,3-葡聚糖酶编码基因(MD11G1189000); (D) 多酚氧化酶编码基因(MD10G1299100); (E) MLP423编码基因(MD16G1088600)。R: 抗病品种华月; S: 感病品种金冠; 不同小写字母表示抗病与感病品种间差异显著(P<0.05)。
Figure 5 Quantitative real-time PCR analysis of genes encoding key resistance-related proteins in apple leaves resistance and susceptible to ring rot disease (A) A gene encoding a thaumatin (MD04G1018400); (B) The gene encoding Mal d1 (MD16G1160700); (C) A gene encoding a beta-1,3-glucosidase (MD11G1189000); (D) A gene encoding a polyphenol oxidase (MD10G1299100); (E) The gene encoding MLP423 (MD16G1088600). R: Resistant cultivars Huayue; S: Susceptible cultivars Golden Delicious; Different lowercase letters indicate significant differences between resistant and susceptible cultivars (P<0.05).
| [1] | 林月莉, 黄丽丽, 索朗拉姆, 高小宁, 陈银潮, 康振生 (2011). 苹果轮纹病室内快速评价体系的建立. 植物保护学报 38, 37-41. |
| [2] | 孙天骅, 李佳, 王涛, 牛宁, 徐继忠 (2018). 抗病与感病苹果叶片应答轮纹病菌侵染的蛋白质表达差异分析. 园艺学报 45, 409-420. |
| [3] | 肖龙, 张彩霞, 宗泽冉, 田义, 张利义, 丛佩华 (2016). 苹果叶片应答轮纹病菌胁迫的叶绿体蛋白质组学分析. 果树学报 33, 1357-1366. |
| [4] | 徐鹏飞, 王萍, 吴俊江, 张淑珍, 范素杰, 李宁辉, 王欣, 陈晨, 李文滨 (2010). 疫霉根腐病菌毒素对大豆不同组织中多酚氧化酶的影响. 作物杂志 ( 2), 28-31. |
| [5] | 杨丽丽, 庄艳, 王忆, 张新忠, 韩振海 (2012). 不同抗性苹果果实受轮纹病菌侵染后亚显微结构的变化. 园艺学报 39, 963-969. |
| [6] | 杨振英, 康国栋, 王强, 薛光荣, 张利义, 田义, 杨玲, 张彩霞, 李武兴, 丛佩华 (2010). 黄色苹果新品种‘华月’. 园艺学报 37, 1877-1878. |
| [7] | 张彩霞, 田义, 张利义, 肖龙, 康国栋, 丛佩华 (2015). 苹果枝条表皮应答轮纹病菌侵染的蛋白质组学分析. 植物病理学报 45, 280-287. |
| [8] | 张彩霞, 袁高鹏, 韩晓蕾, 田义, 张利义, 丛佩华 (2018). 基于iTRAQ定量蛋白质组技术筛选‘华月’苹果斑点落叶病抗性相关蛋白. 植物病理学报 48, 787-798. |
| [9] | 张计育, 渠慎春, 薛华柏, 高志红, 郭忠仁, 章镇 (2012). 北海棠病程相关蛋白MhPR8基因的克隆与表达. 中国农业科学 45, 1568-1575. |
| [10] | 张玉经, 王昆, 王忆, 韩振海, 高源, 许雪峰, 张新忠 (2010). 苹果种质资源果实轮纹病抗性的评价. 园艺学报 37, 539-546. |
| [11] | 周增强, 侯珲, 王丽, 朱发亮 (2010). 枝干苹果轮纹病人工接种方法与品种抗性评价. 果树学报 27, 952-955. |
| [12] | 宗泽冉, 田义, 张利义, 韩晓蕾, 张彩霞, 丛佩华 (2017). 抗苹果斑点落叶病基因Mal d1的克隆及功能鉴定. 园艺学报 44, 343-354. |
| [13] | Berger S, Papadopoulos M, Schreiber U, Kaiser W, Roitsch T (2004). Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol Plant 122, 419-428. |
| [14] |
Berger S, Sinha AK, Roitsch T (2007). Plant physiology meets phytopathology: plant primary metabolism and plant pathogen interactions. J Exp Bot 58, 4019-4026.
DOI URL PMID |
| [15] | Borges LL, Santana FA, Castro ISL, Arruda KMA, de Oliveira Ramos HJ, Moreira MA, de Barros EG (2013). Differentially expressed proteins during an incompatible interaction between common bean and the fungus Pseudocercospora griseola. Mol Breed 32, 933-942. |
| [16] |
Breiteneder H, Ebner C (2000). Molecular and biochemical classification of plant-derived food allergens. J Allergy Clin Immunol 106, 27-36.
DOI URL PMID |
| [17] | Chen Y, Zhang C, Cong P (2012a). Dynamics of growth regulators during infection of apple leaves by Alternaria alternata apple pathotype. Australas Plant Pathol 41, 247-253. |
| [18] |
Chen Z, Wang QH, Lin L, Tang Q, Edwards JL, Li SW, Liu SQ (2012b). Comparative evaluation of two isobaric labeling tags, DiART and iTRAQ. Anal Chem 84, 2908-2915.
DOI URL PMID |
| [19] |
Christensen JH, Bauw G, Welinder KG, van Montagu M, Boerjan M (1998). Purification and characterization of peroxidases correlated with lignification in poplar xylem. Plant Physiol 118, 125-135.
DOI URL PMID |
| [20] |
Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SMN, Baier M, Finkemeier I (2006). The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot 57, 1697-1709.
DOI URL PMID |
| [21] |
Dinakar C, Djilianov D, Bartels D (2012). Photosynthesis in desiccation tolerant plants: energy metabolism and antioxidative stress defense. Plant Sci 182, 29-41.
URL PMID |
| [22] | Eccher G, Ferrero S, Populin F, Colombo L, Botton A (2014). Apple(Malus domestica L. Borkh) as an emerging model for fruit development. Plant Biosyst 148, 157-168. |
| [23] |
Erickson BK, Jedrychowski MP, Mcalister GC, Everley RA, Kunz R, Gygi SP (2015). Evaluating multiplexed quantitative phosphopeptide analysis on a hybrid quadrupole mass filter/linear ion trap/orbitrap mass spectrometer. Anal Chem 87, 1241-1249.
DOI URL PMID |
| [24] | Fan HK, Wang F, Gao H, Wang LC, Xu JH, Zhao ZY (2011). Pathogen-induced MdWRKY1 in ‘Qinguan’ apple enhances disease resistance. J Plant Biol 54, 150-158. |
| [25] | Golba B, Treutter D, Kollar A (2012). Effects of apple (Malus × domestica Borkh.) phenolic compounds on proteins and cell wall-degrading enzymes of Venturia inaequalis. Trees 26, 131-139. |
| [26] | Grover A (2012). Plant chitinases: genetic diversity and physiological roles. Crit Rev Plant Sci 31, 57-73. |
| [27] | Jurick II WM, Janisiewicz WJ, Saftner RA, Vico I, Gaskins VL, Park E, Forsline PL, Fazio G, Conway WS (2011). Identification of wild apple germplasm(Malus spp.) accessions with resistance to the postharvest decay pathogens Penicillium expansum and Colletotrichum acutatum. Plant Breed 130, 481-486. |
| [28] | Kim BG, Fukumoto T, Tatano S, Gomi K, Ohtani K, Tada Y, Akimitsu K (2009). Molecular cloning and characterization of a thaumatin-like protein-encoding cDNA from rough lemon. Physiol Mol Plant Pathol 74, 3-10. |
| [29] |
Kornas A, Kuźniak E, Slesak I, Miszalski I (2010). The key role of the redox status in regulation of metabolism in photosynthesizing organisms. Acta Biochim Pol 57, 143-151.
URL PMID |
| [30] |
Li L, Steffens JC (2002). Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta 215, 239-247.
DOI URL PMID |
| [31] |
Li MM, Xu JH, Qiu ZH, Zhang J, Ma FW, Zhang JK (2014). Screening and identification of resistance related proteins from apple leaves inoculated with Marssonina coronaria(EII. & J. J. Davis). Proteome Sci 12, 7.
DOI URL PMID |
| [32] |
Li ZT, Dhekney SA, Grad DJ (2011). PR-1 gene family of grapevine: a uniquely duplicated PR-1 gene from a Vitis interspecific hybrid confers high level resistance to bacterial disease in transgenic tobacco. Plant Cell Rep 30, 1-11.
DOI URL |
| [33] |
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402-408.
DOI URL PMID |
| [34] |
Mazzeo MF, Cacace G, Ferriello F, Puopolo G, Zoina A, Ercolano MR, Siciliano RA (2014). Proteomic investigation of response to forl infection in tomato roots. Plant Physiol Biochem 74, 42-49.
DOI URL PMID |
| [35] |
Ni WC, Zhu LM, Sha RH, Tao JM, Cai BH, Wang SH (2017). Comparative iTRAQ proteomic profiling of susceptible and resistant apple cultivars infected by Alternaria alternata apple pathotype. Tree Genet Genomes 13, 23.
DOI URL |
| [36] |
Ogata T, Sano T, Harada Y (2000). Botryosphaeria spp. isolated from apple and several deciduous fruit trees are divided into three groups based on the production of warts on twigs, size of conidia, and nucleotide sequences of nuclear ribosomal DNA ITS regions. Mycoscience 41, 331-337.
DOI URL |
| [37] |
Paulo JA, McAllister FE, Everley RA, Beausoleil SA, Bank AS, Gygi SP (2015). Effects of MEK inhibitors GSK1120212 and PD0325901 in vivo using 10-plex quantitative proteomics and phosphoproteomics. Proteomics 15, 462-473.
DOI URL PMID |
| [38] |
Ramsubramaniam N, Tao F, Li SW, Marten MR (2013). Cost-effective isobaric tagging for quantitative phosphoproteomics using DiART reagents. Mol Biosyst 9, 2981-2987.
DOI URL |
| [39] |
Savitski MM, Wilhelm M, Hahne H, Kuster B, Bantscheff M (2015). A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol Cell Proteom 14, 2394-2404.
DOI URL |
| [40] |
Sun H, Kim MK, Pulla RK, Kim YJ, Yang DC (2010). Isolation and expression analysis of a novel major latex-like protein (MLP151) gene from Panax ginseng. Mol Biol Rep 37, 2215-2222.
DOI URL PMID |
| [41] |
Thipyapong P, Hunt MD, Steffens JC (2004). Antisense downregulation of polyphenol oxidase results in enhanced disease susceptibility. Planta 220, 105-117.
DOI URL |
| [42] |
Wang YP, Yang L, Chen X, Ye TT, Zhong B, Liu RJ, Wu Y, Chan ZL (2016). Major latex protein-like protein 43 (MLP43) functions as a positive regulator during abscisic acid responses and confers drought tolerance in Arabidopsis thaliana. J Exp Bot 67, 421-434.
DOI URL PMID |
| [43] |
Wen B, Zhou R, Feng Q, Wang QH, Wang J, Liu SQ (2014). IQuant: an automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 14, 2280-2285.
URL PMID |
| [44] |
Xing LL, Sun LN, Liu SL, Li XN, Zhang LB, Yang HS (2017). IBT-based quantitative proteomics identifies potential regulatory proteins involved in pigmentation of purple sea cucumber, Apostichopus japonicus. Comp Biochem Physiol Part D Genomics Proteomics 23, 17-26.
DOI URL PMID |
| [45] | Xu C, Wang CS, Ju LL, Zhang R, Biggs AR, Tanaka E, Li BZ, Sun GY (2015). Multiple locus genealogies and phenotypic characters reappraise the causal agents of apple ring rot in China. Fungal Divers 71, 215-231. |
| [46] |
Zhang CX, Tian Y, Cong PH (2015). Proteome analysis of pathogen-responsive proteins from apple leaves induced by the Alternaria blotch Alternaria alternata. PLoS One 10, e0122233.
DOI URL PMID |
| [1] | 王秀媛, 申磊, 刘婷婷, 尉雯雯, 张帅, 张伟. ‘塞外红’苹果-大豆复合系统根系时空分布与种间竞争策略[J]. 植物生态学报, 2025, 49(5): 748-759. |
| [2] | 路晨曦, 徐漫, 石学瑾, 赵成, 陶泽, 李敏, 司炳成. 基于贝叶斯模型MixSIAR的不同水同位素输入方法对苹果园吸水特征分析结果的影响[J]. 植物生态学报, 2023, 47(2): 238-248. |
| [3] | 张宏祥, 闻志彬, 王茜. 新疆野苹果种群遗传结构及其环境适应性[J]. 植物生态学报, 2022, 46(9): 1098-1108. |
| [4] | 张娜,刘秀霞,陈学森,吴树敬. 基于转录组分析鉴定苹果茉莉素响应基因[J]. 植物学报, 2019, 54(6): 733-743. |
| [5] | 何光明, 邓兴旺. 死亡信号传递: 叶绿体与线粒体间信号交流调控植物程序性细胞死亡[J]. 植物学报, 2018, 53(4): 441-444. |
| [6] | 赵曦娟, 钱礼超, 刘玉乐. 中国科学家在植物程序性细胞死亡领域取得重要成果[J]. 植物学报, 2018, 53(4): 447-450. |
| [7] | 张宪省. 我国科学家在程序性细胞死亡机制研究领域取得重大突破[J]. 植物学报, 2018, 53(4): 445-446. |
| [8] | 张彩霞, 田义, 张利义, 肖龙, 丛佩华. 苹果属植物树皮组织总蛋白提取及分离方法的建立[J]. 植物学报, 2015, 50(6): 739-745. |
| [9] | 杨启良, 张富仓, 刘小刚, 杨振宇. 不同滴灌方式和NaCl处理对苹果幼树生长和水分传导的影响[J]. 植物生态学报, 2009, 33(4): 824-832. |
| [10] | 田义;王强;张利义;康国栋;杨玲;郝红梅;杨振英;丛佩华*. 外源腐胺促进苹果果皮花青苷积累的效应[J]. 植物学报, 2009, 44(03): 310-316. |
| [11] | 田义;张新忠;张志宏;丛佩华;康国栋. 无胶筛分毛细管电泳在分析苹果发育时期转变蛋白质变化规律中的应用[J]. 植物学报, 2008, 25(05): 585-590. |
| [12] | 张光灿, 刘霞, 贺康宁, 王百田. 金矮生苹果叶片气体交换参数对土壤水分的响应[J]. 植物生态学报, 2004, 28(1): 66-72. |
| [13] | 雷晓勇 黄蕾 田梅生 胡小松 戴尧仁. 苹果果肉中抗氰氧化酶(AOX)的分离鉴定[J]. 植物学报, 2002, 19(06): 739-742. |
| [14] | 周志钦, 成明昊, 宋洪元, 李晓林, 杨天秀. 苹果属小金海棠的遗传多样性初步研究[J]. 生物多样性, 2001, 09(2): 145-150. |
| [15] | 关军锋. ca2 +对苹果果实细胞膜透性 保护酶活性和保护物质含量的影响[J]. 植物学报, 1999, 16(01): 72-74. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||