

植物学报 ›› 2018, Vol. 53 ›› Issue (3): 305-312.DOI: 10.11983/CBB18027 cstr: 32102.14.CBB18027
所属专题: 药用植物专辑 (2018年53卷3期)
朱宇佳1, 焦凯丽1, 罗秀俊1, 冯尚国1,2,*( ), 王慧中1,2,*(
), 王慧中1,2,*( )
)
收稿日期:2018-01-28
接受日期:2018-05-09
出版日期:2018-05-01
发布日期:2018-06-05
通讯作者:
冯尚国,王慧中
基金资助:
Zhu Yujia1, Jiao Kaili1, Luo Xiujun1, Feng Shangguo1,2,*( ), Wang Huizhong1,2,*(
), Wang Huizhong1,2,*( )
)
Received:2018-01-28
Accepted:2018-05-09
Online:2018-05-01
Published:2018-06-05
Contact:
Feng Shangguo,Wang Huizhong
摘要: 近年来, 由于营养价值高、果实可当水果食用且具有潜在的药用价值, 酸浆属(Physalis)植物在全球范围内受到了越来越多的关注。采用SSR分子标记技术对我国范围内主要分布的4种酸浆属植物的22份样品进行了亲缘关系研究, 结果表明, 20对SSR引物共扩增出118个位点, 其中107个(90.7%)为多态性位点; 平均种间遗传相似系数为0.501。UPGMA聚类和PCoA分析结果得出相似结论, 并且将供试酸浆属植物样品分为两大类。其中, 酸浆(P. alkekengi var. francheti)的所有样品与其它酸浆属植物的遗传距离最远, 单独聚为一类, 与前人的研究结果非常吻合。研究表明, SSR标记遗传信息丰富, 可以用于酸浆属植物的亲缘关系分析。研究结果为酸浆属种质资源保护提供了有效信息, 并且为酸浆属植物的分子辅助育种奠定了重要基础。
朱宇佳, 焦凯丽, 罗秀俊, 冯尚国, 王慧中. 基于SSR分子标记的酸浆属植物亲缘关系研究. 植物学报, 2018, 53(3): 305-312.
Zhu Yujia, Jiao Kaili, Luo Xiujun, Feng Shangguo, Wang Huizhong. Genetic Relationship of Physalis Plants Revealed by Simple Sequence Repeat Markers. Chinese Bulletin of Botany, 2018, 53(3): 305-312.
| No. | Species name | Code | Origin |
|---|---|---|---|
| 1 | Physalis minima | XSJ1 | Qianxi, Tangshan, Hebei |
| 2 | P. minima | XSJ2 | Mudan, Heze, Shandong |
| 3 | P. minima | XSJ3 | Lou’an, Anhui |
| 4 | P. minima | XSJ4 | Lishui, Zhejiang |
| 5 | P. angulata | KZ1 | Jianggan, Hangzhou, Zhejiang |
| 6 | P. angulata | KZ2 | Xiaoshan, Hangzhou, Zhejiang |
| 7 | P. angulata | KZ3 | Luotian, Huanggang, Hubei |
| 8 | P. angulata | KZ4 | Honghe, Yunnan |
| 9 | P. angulata | KZ5 | Najing, Jiangsu |
| 10 | P. angulata | KZ6 | Linhai, Taizhou, Zhejiang |
| 11 | P. angulata | KZ7 | Wenzhou, Zhejiang |
| 12 | P. angulata | KZ8 | Pujiang, Jinhua, Zhejiang |
| 13 | P. angulata | KZ9 | Xiajin, Dezhou, Shandong |
| 14 | P. alkekengi var. francheti | SJ1 | Faku, Shenyang, Liaoning |
| 15 | P. alkekengi var. francheti | SJ2 | Donggang, Dandong, Liaoning |
| 16 | P. alkekengi var. francheti | SJ3 | Nong’an, Changchun, Jilin |
| 17 | P. alkekengi var. francheti | SJ4 | Zoucheng, Jinan, Shandong |
| 18 | P. pubescens | MSJ1 | Faku, Shenyang, Liaoning |
| 19 | P. pubescens | MSJ2 | Nong’an, Changchun, Jilin |
| 20 | P. pubescens | MSJ3 | Chaoyang, Zhaodong, Heilongjiang |
| 21 | P. pubescens | MSJ4 | Mudanjiang, Heilongjiang |
| 22 | P. pubescens | MSJ5 | Hulunbeir, Inner Mongolia |
表1 供试的22份酸浆属植物样本材料
Table 1 Twenty-two Physalis samples tested in this experiment
| No. | Species name | Code | Origin |
|---|---|---|---|
| 1 | Physalis minima | XSJ1 | Qianxi, Tangshan, Hebei |
| 2 | P. minima | XSJ2 | Mudan, Heze, Shandong |
| 3 | P. minima | XSJ3 | Lou’an, Anhui |
| 4 | P. minima | XSJ4 | Lishui, Zhejiang |
| 5 | P. angulata | KZ1 | Jianggan, Hangzhou, Zhejiang |
| 6 | P. angulata | KZ2 | Xiaoshan, Hangzhou, Zhejiang |
| 7 | P. angulata | KZ3 | Luotian, Huanggang, Hubei |
| 8 | P. angulata | KZ4 | Honghe, Yunnan |
| 9 | P. angulata | KZ5 | Najing, Jiangsu |
| 10 | P. angulata | KZ6 | Linhai, Taizhou, Zhejiang |
| 11 | P. angulata | KZ7 | Wenzhou, Zhejiang |
| 12 | P. angulata | KZ8 | Pujiang, Jinhua, Zhejiang |
| 13 | P. angulata | KZ9 | Xiajin, Dezhou, Shandong |
| 14 | P. alkekengi var. francheti | SJ1 | Faku, Shenyang, Liaoning |
| 15 | P. alkekengi var. francheti | SJ2 | Donggang, Dandong, Liaoning |
| 16 | P. alkekengi var. francheti | SJ3 | Nong’an, Changchun, Jilin |
| 17 | P. alkekengi var. francheti | SJ4 | Zoucheng, Jinan, Shandong |
| 18 | P. pubescens | MSJ1 | Faku, Shenyang, Liaoning |
| 19 | P. pubescens | MSJ2 | Nong’an, Changchun, Jilin |
| 20 | P. pubescens | MSJ3 | Chaoyang, Zhaodong, Heilongjiang |
| 21 | P. pubescens | MSJ4 | Mudanjiang, Heilongjiang |
| 22 | P. pubescens | MSJ5 | Hulunbeir, Inner Mongolia |
| Primer name | Primer sequence (5'-3') | Repeat type | Tm | No. of loci | Polymorphic loci | Polymorphism rate (%) |
|---|---|---|---|---|---|---|
| SSR2 | F: CATTGGGTTTCGCATCCAT | AG | 60 | 6 | 6 | 100.0 |
| R: AGACAAGCCTAGGGGAAAGG | ||||||
| SSR9 | F: TGCTCCGAGTTTTAGGGTTC | AG | 60 | 8 | 7 | 87.5 |
| R: GCAGTTGGTAAAGTTGAGAGACG | ||||||
| SSR10 | F: GCTTCCTATTGTGTTGCCTGA | AT | 58 | 5 | 4 | 80.0 |
| R: ACTTTGGGTTTCGGGAATTG | ||||||
| SSR11 | F: CAGCTGAAATAAGAGAGTGATTGG | AG | 57 | 4 | 3 | 75.0 |
| R: CCCTCTTTTTCTCCTCCGAGT | ||||||
| SSR13 | F: GCGGAATCCATTGTTTTTCA | AC | 58 | 9 | 8 | 88.9 |
| R: CCGATGGAGTATAGTCACGCAAA | ||||||
| SSR15 | F: GCTTGTTGATCAGCTTTCTTTG | AT | 57 | 7 | 6 | 85.7 |
| R: TGGATCATAACCTTGCTAATGC | ||||||
| SSR36 | F: ATGAACCACATGTCGGAGGA | AG | 58 | 7 | 6 | 85.7 |
| R: GGGGATCCAAACGAAGTGTA | ||||||
| SSR54 | F: CGGCTGGTATGCTTACAAAGAT | AC | 58 | 4 | 4 | 100.0 |
| R: GCACTTCCACTGTTTTTAACTTCC | ||||||
| SSR55 | F: CACCTACATAGGCAGCCAAAA | AG | 58 | 7 | 6 | 85.7 |
| R: ATTTGTGGGCGGAGGAAG | ||||||
| SSR57 | F: AGTGAAAAGCAGCCCATTCT | AT | 56 | 9 | 8 | 88.9 |
| R: GGCGAAGCTGAATTGAAAAA | ||||||
| SSR67 | F: GCTTCTGTTCCATTATTCACCA | AG | 56 | 5 | 5 | 100.0 |
| R: GCAGTGTGGGATCAATCAAT | ||||||
| SSR68 | F: GAAGCAAACAACTACACCCAAA | AG | 56 | 8 | 8 | 100.0 |
| R: AAGCCTCGGATTTCATAGCA | ||||||
| SSR77 | F: CATACCATAACTCCCCATCTCTC | AG | 57 | 4 | 3 | 75.0 |
| R: TGCCGATTCTGATTTCTTCC | ||||||
| SSR92 | F: TGGTTTGAGGATCAAGAAAGAA | AAG | 56 | 5 | 4 | 80.0 |
| R: GTGGTATCAACGCAGAGTGG | ||||||
| SSR107 | F: CATCCAACACCAGAAATACGC | AAG | 58 | 4 | 4 | 100.0 |
| R: TCCAACTTTATCATTTCTTCCAC | ||||||
| SSR110 | F: CACCCATATCCCAATCTTCTTC | CTT | 60 | 4 | 4 | 100.0 |
| R: GGGTAATTTTCACGGGGAAT | ||||||
| SSR112 | F: CTACGCCTACCACTTGCACA | TCT | 60 | 11 | 11 | 100.0 |
| R: CAGTGGAAGCCTCAAGATCC | ||||||
| SSR118 | F: AATCAAGGGTCAGAAGAAATGG | ATC | 58 | 2 | 2 | 100.0 |
| R: GCAAGAATGGATGTGGGTGT | ||||||
| SSR123 | F: TCAGTGGAGCGCGTATATCT | ATC | 60 | 5 | 5 | 100.0 |
| R: GCGATCTCACCAAACCTCTC | ||||||
| SSR127 | F: TTGGTTTGGCATAACTGCAA | AAT | 58 | 4 | 3 | 75.0 |
| R: GGTTTGCAACTCTCATGCTG | ||||||
| Average | - | - | - | 5.9 | 5.4 | 90.4 |
| Total | - | - | - | 118 | 107 | - |
表2 20对简单重复序列(SSR)引物的扩增结果及多态性信息
Table 2 Amplification results and polymorphism information of 20 simple sequence repeats (SSR) primer pairs
| Primer name | Primer sequence (5'-3') | Repeat type | Tm | No. of loci | Polymorphic loci | Polymorphism rate (%) |
|---|---|---|---|---|---|---|
| SSR2 | F: CATTGGGTTTCGCATCCAT | AG | 60 | 6 | 6 | 100.0 |
| R: AGACAAGCCTAGGGGAAAGG | ||||||
| SSR9 | F: TGCTCCGAGTTTTAGGGTTC | AG | 60 | 8 | 7 | 87.5 |
| R: GCAGTTGGTAAAGTTGAGAGACG | ||||||
| SSR10 | F: GCTTCCTATTGTGTTGCCTGA | AT | 58 | 5 | 4 | 80.0 |
| R: ACTTTGGGTTTCGGGAATTG | ||||||
| SSR11 | F: CAGCTGAAATAAGAGAGTGATTGG | AG | 57 | 4 | 3 | 75.0 |
| R: CCCTCTTTTTCTCCTCCGAGT | ||||||
| SSR13 | F: GCGGAATCCATTGTTTTTCA | AC | 58 | 9 | 8 | 88.9 |
| R: CCGATGGAGTATAGTCACGCAAA | ||||||
| SSR15 | F: GCTTGTTGATCAGCTTTCTTTG | AT | 57 | 7 | 6 | 85.7 |
| R: TGGATCATAACCTTGCTAATGC | ||||||
| SSR36 | F: ATGAACCACATGTCGGAGGA | AG | 58 | 7 | 6 | 85.7 |
| R: GGGGATCCAAACGAAGTGTA | ||||||
| SSR54 | F: CGGCTGGTATGCTTACAAAGAT | AC | 58 | 4 | 4 | 100.0 |
| R: GCACTTCCACTGTTTTTAACTTCC | ||||||
| SSR55 | F: CACCTACATAGGCAGCCAAAA | AG | 58 | 7 | 6 | 85.7 |
| R: ATTTGTGGGCGGAGGAAG | ||||||
| SSR57 | F: AGTGAAAAGCAGCCCATTCT | AT | 56 | 9 | 8 | 88.9 |
| R: GGCGAAGCTGAATTGAAAAA | ||||||
| SSR67 | F: GCTTCTGTTCCATTATTCACCA | AG | 56 | 5 | 5 | 100.0 |
| R: GCAGTGTGGGATCAATCAAT | ||||||
| SSR68 | F: GAAGCAAACAACTACACCCAAA | AG | 56 | 8 | 8 | 100.0 |
| R: AAGCCTCGGATTTCATAGCA | ||||||
| SSR77 | F: CATACCATAACTCCCCATCTCTC | AG | 57 | 4 | 3 | 75.0 |
| R: TGCCGATTCTGATTTCTTCC | ||||||
| SSR92 | F: TGGTTTGAGGATCAAGAAAGAA | AAG | 56 | 5 | 4 | 80.0 |
| R: GTGGTATCAACGCAGAGTGG | ||||||
| SSR107 | F: CATCCAACACCAGAAATACGC | AAG | 58 | 4 | 4 | 100.0 |
| R: TCCAACTTTATCATTTCTTCCAC | ||||||
| SSR110 | F: CACCCATATCCCAATCTTCTTC | CTT | 60 | 4 | 4 | 100.0 |
| R: GGGTAATTTTCACGGGGAAT | ||||||
| SSR112 | F: CTACGCCTACCACTTGCACA | TCT | 60 | 11 | 11 | 100.0 |
| R: CAGTGGAAGCCTCAAGATCC | ||||||
| SSR118 | F: AATCAAGGGTCAGAAGAAATGG | ATC | 58 | 2 | 2 | 100.0 |
| R: GCAAGAATGGATGTGGGTGT | ||||||
| SSR123 | F: TCAGTGGAGCGCGTATATCT | ATC | 60 | 5 | 5 | 100.0 |
| R: GCGATCTCACCAAACCTCTC | ||||||
| SSR127 | F: TTGGTTTGGCATAACTGCAA | AAT | 58 | 4 | 3 | 75.0 |
| R: GGTTTGCAACTCTCATGCTG | ||||||
| Average | - | - | - | 5.9 | 5.4 | 90.4 |
| Total | - | - | - | 118 | 107 | - |
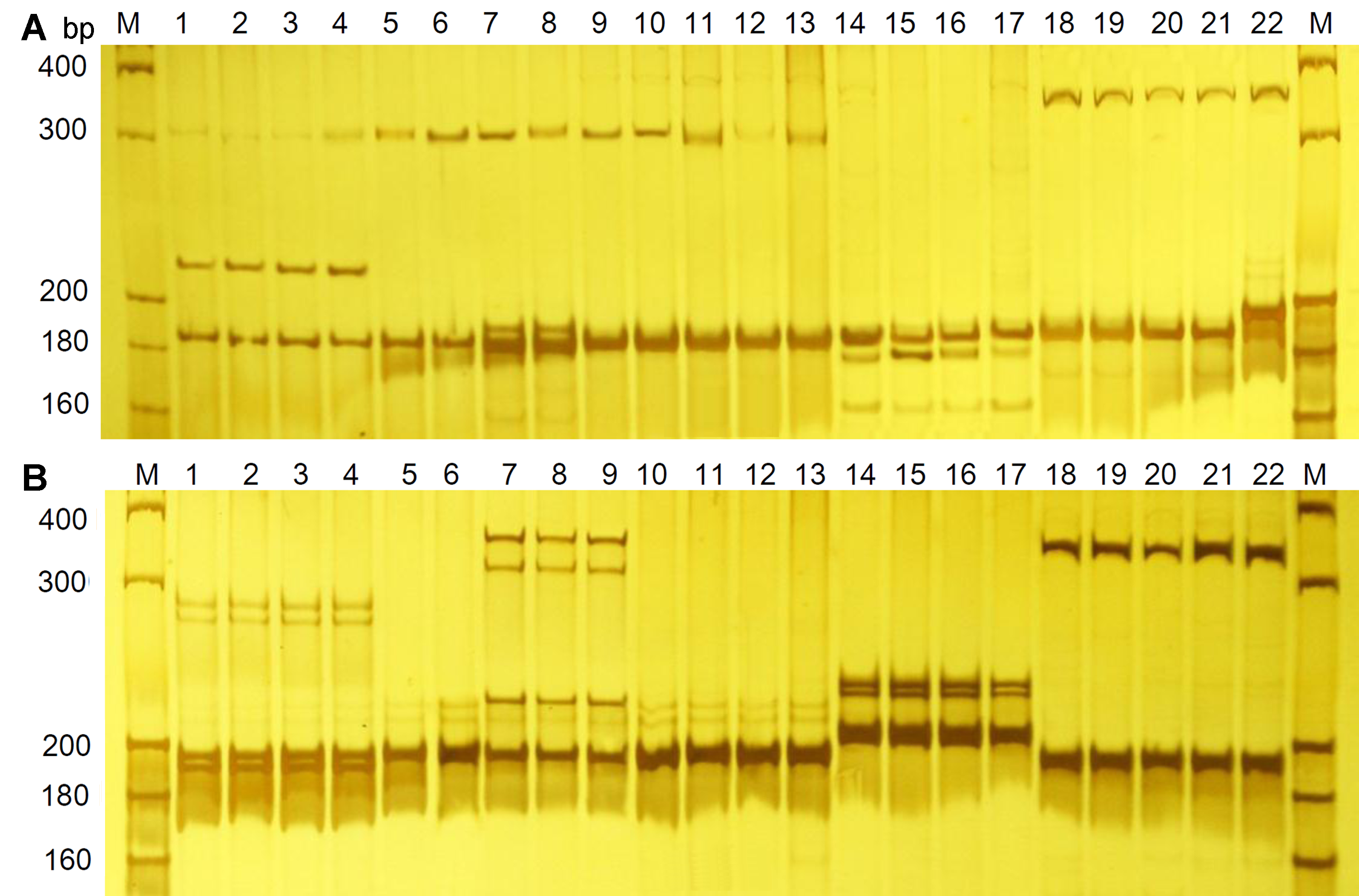
图1 引物SSR55 (A)和SSR112 (B)在酸浆属植物中的电泳检测结果M: DNA分子标准。泳道1-22: 22个酸浆属样品(编号同表1)
Figure 1 Amplification profile of primer SSR55 (A) and SSR112 (B) in Physalis samplesM: DNA molecular standards. Lane 1-22: The 22 Physalis samples (sample number is the same as in Table 1).
| Measurement | Similarity coefficient |
|---|---|
| Average interspecies genetic similarity coefficient | 0.501±0.074 |
| P. angulata vs P. minima | 0.600±0.042 |
| P. alkekengi var. francheti vs P. minima | 0.437±0.036 |
| P. pubescens vs P. minima | 0.570±0.037 |
| P. angulata vs P. alkekengi var. francheti | 0.444±0.044 |
| P. angulata vs P. pubescens | 0.514±0.043 |
| P. pubescens vs P. alkekengi var. francheti | 0.382±0.040 |
表3 酸浆属植物平均种间遗传相似系数
Table 3 Average interspecies genetic similarity coefficient in Physalis samples
| Measurement | Similarity coefficient |
|---|---|
| Average interspecies genetic similarity coefficient | 0.501±0.074 |
| P. angulata vs P. minima | 0.600±0.042 |
| P. alkekengi var. francheti vs P. minima | 0.437±0.036 |
| P. pubescens vs P. minima | 0.570±0.037 |
| P. angulata vs P. alkekengi var. francheti | 0.444±0.044 |
| P. angulata vs P. pubescens | 0.514±0.043 |
| P. pubescens vs P. alkekengi var. francheti | 0.382±0.040 |
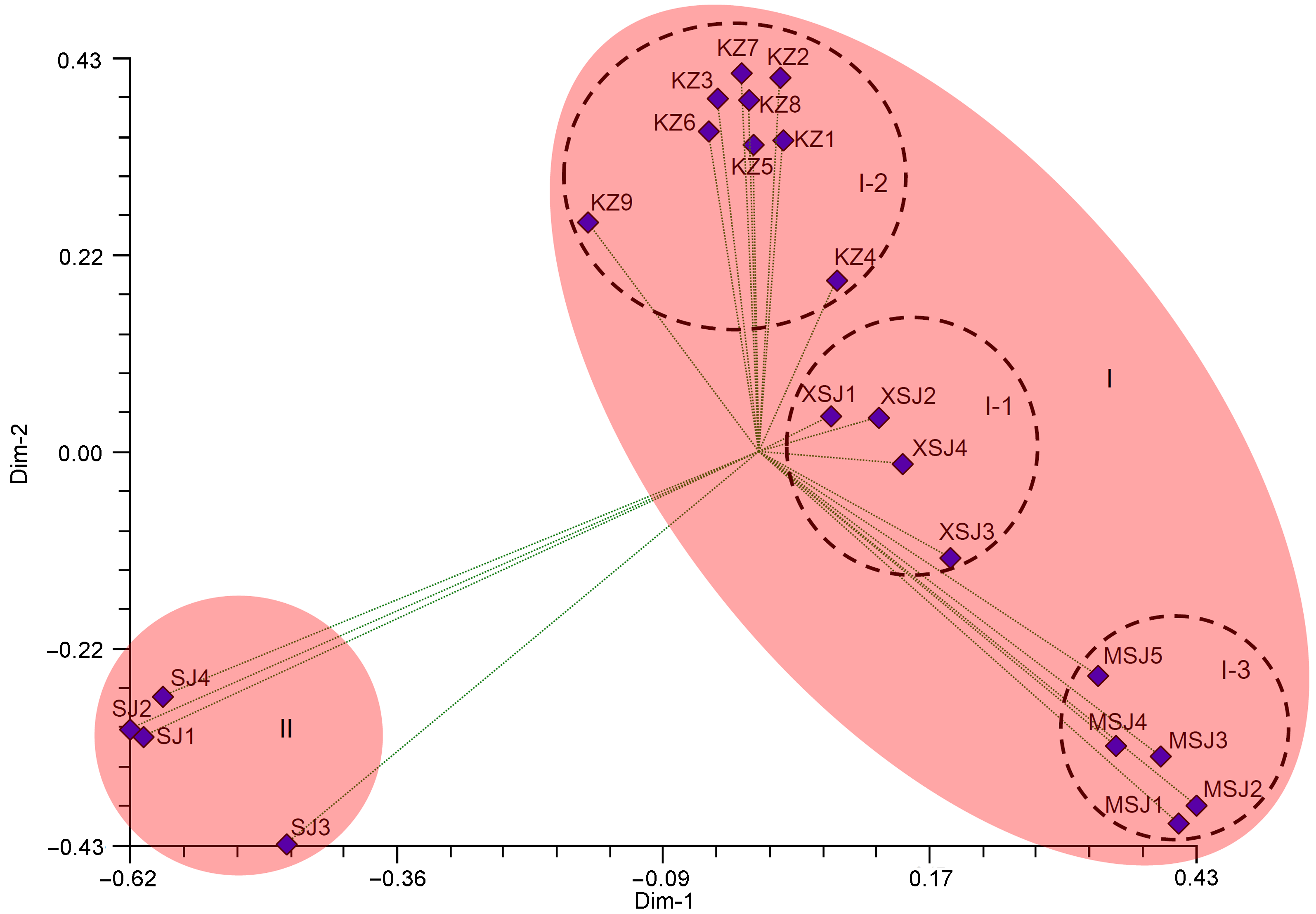
图3 基于简单重复序列(SSR)标记的22份酸浆属植物样品二维主坐标分析编码同表1。
Figure 3 The analysis of PCoA for 22 Physalis samples based on simple sequence repeats (SSR) markers along the first two principal axesThe codes are the same as in Table 1.
| [1] | 国家药典委员会 (2015). 中华人民共和国药典(一部). 北京: 中国医药科技出版社. pp. 360-361. |
| [2] | 郭瑜, 吴东栋, 任志远, 王晓闻, 郭峰 (2017). 酸浆悬浮饮料的研制. 食品研究与开发 38(15), 89-92. |
| [3] | 贾远敏, 陈重, 许琼明, 李笑然, 杨世林 (2013). 毛酸浆浆果的化学成分研究. 中草药 9, 1086-1090. |
| [4] | 李晓岚, 陆嘉惠, 谢良碧, 张爱霞, 陈晓翠, 李学禹 (2015). 4种甘草属植物EST-SSR引物开发及其亲缘关系分析. 西北植物学报 35, 480-485. |
| [5] | 骆丽萍, 成凡钦, 季龙, 虞和永 (2015). 毛酸浆的化学成分研究. 中国中药杂志 22, 4424-4427. |
| [6] | 孙海涛, 高玉超 (2013). 毛酸浆番茄复合调味酱的研制. 中国调味品 38(12), 58-59, 67. |
| [7] | 王晓英, 刘长姣, 段连海, 霍岩 (2014). 毛酸浆开发利用的研究进展. 中国酿造 2, 5-8. |
| [8] | 王赢, 朱丹, 牛广财, 郑唯, 魏文毅, 刘鑫 (2017). 毛酸浆发酵果脯的研制. 中国酿造 36(8), 182-185. |
| [9] | 许亮, 王荣祥, 杨燕云, 王冰 (2009). 中国酸浆属植物药用资源研究. 中国野生植物资源 28, 21-23. |
| [10] | 杨金颖, 杨德草, 李津津 (2014). 酸浆的药理作用研究进展. 内蒙古中医药 33(25), 116-117. |
| [11] | 中国科学院中国植物志编辑委员会 (1978). 中国植物志(第67卷). 北京: 科学出版社. pp. 50. |
| [12] | Axelius B (1996). The phylogenetic relationships of the physaloid genera (Solanaceae) based on morphological data.Am J Bot 83, 118-124. |
| [13] | Ding H, Hu ZJ, Yu LY, Ma ZY, Ma XQ, Chen Z, Wang D, Zhao XF (2014). Induction of quinone reductase (QR) by withanolides isolated from Physalis angulata L. var. villosa Bonati (Solanaceae). Steroids 86, 32-38. |
| [14] | Feng SG, He RF, Lu JJ, Jiang MY, Shen XY, Jiang Y, Wang ZA, Wang HZ (2016a). Development of SSR markers and assessment of genetic diversity in medicinal Chrysanthemum morifolium cultivars. Front Genet 7, 113. |
| [15] | Feng SG, Jiang MY, Shi YJ, Jiao KL, Shen CJ, Lu JJ, Ying QC, Wang HZ (2016b). Application of the ribosomal DNA ITS2 region of Physalis ( Solanaceae): DNA barcoding and phylogenetic study. Front Plant Sci 7, 1047. |
| [16] | Feng SG, Jiao KL, Zhu YJ, Wang HF, Jiang MY, Wang HZ (2018). Molecular identification of species of Physalis ( Solanaceae) using a candidate DNA barcode: the chloroplast psbA-trnH intergenic region. Genome 61, 15-20. |
| [17] | Fu N, Wang PY, Liu XD, Shen HL (2014). Use of EST-SSR markers for evaluating genetic diversity and fingerprinting celery (Apium graveolens L.) cultivars. Molecules 19, 1939-1955. |
| [18] | Garzon-Martínez GA, Osorio-Guarín JA, Delgádillo-Duran P, Mayorga F, Enciso-Rodríguez FE, Landsman D, Mariño-Ramírez L, Barrero LS (2015). Genetic diversity and population structure in Physalis peruviana and related taxa based on InDels and SNPs derived from COSII and IRG markers. Plant Gene 4, 29-37. |
| [19] | Gower JC (1966). Some distance properties of latent root and vector methods used in multivariate analysis.Biometrika 53, 325-338. |
| [20] | Ji L, Yuan YL, Ma ZJ, Chen Z, Gan LS, Ma XQ, Huang DS (2013). Induction of quinone reductase (QR) by withanolides isolated from Physalis pubescens L. ( Solanaceae). Steroids 78, 860-865. |
| [21] | Li X, Zhao JP, Yang M, Liu YL, Li ZC, Li RY, Li XR, Li N, Xu QM, Khan IA, Yang SL (2014). Physalins and withanolides from the fruits ofPhysalis alkekengi L. var. fran- chetii ( Mast.) Makino and the inhibitory activities against human tumor cells. Phytochem Lett 10, 95-100. |
| [22] | Maggie WPS, Manos PS (2005). Untangling Physalis ( Solanaceae) from the physaloids: a two-gene phylogeny of the Physalinae. Syst Bot 30, 216-230. |
| [23] | Nei M, Li WH (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases.Proc Natl Acad Sci USA 76, 5269-5273. |
| [24] | Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, Collier SM (2008). A molecular phylogeny of the Solanaceae.Taxon 57, 1159-1181. |
| [25] | Rohlf FJ (2000). NTSYS-PC: numerical taxonomy and multivariate analysis system, version 2.00. Setauket, New York:Exeter Software. |
| [26] | Simbaqueba J, Sánchez P, Sanchez E, Zarantes VMN, Chacon MI, Barrero LS, Mariño-Ramírez L (2011). Development and characterization of microsatellite markers for the cape gooseberry Physalis peruviana. PLoS One 6, e26719. |
| [27] | Sun CP, Nie XF, Kang N, Zhao F, Chen LX, Qiu F (2017). A new phenol glycoside from Physalis angulata. Nat Prod Res 31, 1059-1065. |
| [28] | Teshome A, Bryngelsson T, Dagne K, Geleta M (2015). Assessment of genetic diversity in Ethiopian field pea ( Pisum sativum L.) accessions with newly developed EST- SSR markers. BMC Genet 16, 102. |
| [29] | Wang F, Yang T, Burlyaeva M, Li L, Jiang JY, Fang L, Redden R, Zong XX (2015). Genetic diversity of grasspea and its relative species revealed by SSR markers.PLoS One 10, e0118542. |
| [30] | Wei JL, Hu XR, Yang JJ, Yang WC (2012). Identification of single-copy orthologous genes between Physalis and Solanum lycopersicum and analysis of genetic diversity in Physalis using molecular markers. PLoS One 7, e50164. |
| [31] | Xu XM, Guan YZ, Shan SM, Luo JG, Kong LY (2016). Withaphysalin-type withanolides from Physalis minima. Phytochem Lett 15, 1-6. |
| [32] | Yang YK, Xie SD, Xu WX, Nian Y, Liu XL, Peng XR, Ding ZT, Qiu MH (2016). Six new physalins from Physalis alkekengi var. franchetii and their cytotoxicity and antibacterial activity. Fitoterapia 112, 144-152. |
| [33] | Zamora-Tavares P, Vargas-Ponce O, Sanchez-Martínez J, Cabrera-Toledo D (2015). Diversity and genetic structure of the husk tomato ( Physalis philadelphica Lam.) in Wes- tern Mexico. Genet Res Crop Evol 62, 141-153. |
| [34] | Zhang WN, Tong WY (2016). Chemical constituents and biological activities of plants from the genus Physalis. Chem Biodivers 13, 48-65. |
| [1] | 陈旭, 王国严, 彭培好, 李景吉, 石松林, 张廷斌. 四川攀西地区云南松群落物种多样性和谱系多样性对紫茎泽兰入侵的影响[J]. 生物多样性, 2021, 29(7): 865-874. |
| [2] | 赵颖, 马荣, 尹永香, 张志东, 田呈明. 新疆不同来源金黄壳囊孢的多样性[J]. 生物多样性, 2019, 27(10): 1122-1131. |
| [3] | 胡亚妮, 张宗文, 吴斌, 高佳, 李艳琴. 基于ITS和ndhF-rpl32序列的荞麦种间亲缘关系分析[J]. 生物多样性, 2016, 24(3): 296-303. |
| [4] | 郭琪, 郭大龙, 郭丽丽, 张琳, 侯小改. SSR分子标记在牡丹亲缘关系研究中的应用与研究进展[J]. 植物学报, 2015, 50(5): 652-664. |
| [5] | 刘文, 梁红, 杨碧忍, 林杰文, 胡延吉. 一种高等植物小片段RNA分子标记方法[J]. 生物多样性, 2012, 20(1): 86-93. |
| [6] | 罗向东, 戴亮芳, 万勇, 胡标林, 李佛生, 李霞, 谢建坤. 东乡野生稻与栽培稻正反交种间杂种F1的雄配子发生与发育[J]. 植物学报, 2011, 46(4): 407-412. |
| [7] | 陈英. 常绿阔叶林谱系多样性对幼苗存活率的影响[J]. 植物生态学报, 2009, 33(6): 1084-1089. |
| [8] | 严茂粉, 李向华, 王克晶. 北京地区野生大豆种群SSR标记的遗传多样性评价[J]. 植物生态学报, 2008, 32(4): 938-950. |
| [9] | 李潞滨;郭晓军;彭镇华;刘贯水;袁洪水;朱宝成;杨凯*. AFLP 引物组合数量对准确研究竹子系统关系的影响[J]. 植物学报, 2008, 25(04): 449-454. |
| [10] | 王丽侠, 李英慧, 李伟, 朱莉, 关媛, 宁学成, 关荣霞, 刘章雄, 常汝镇, 邱丽娟. 长江春大豆核心种质构建及分析[J]. 生物多样性, 2004, 12(6): 578-585. |
| [11] | 吴萍, 李奕仁, 王金玉, 陈宽维. 应用微卫星标记分析中国地方鸡种的遗传变异[J]. 生物多样性, 2003, 11(6): 461-466. |
| [12] | 李太武, 李成华, 宋林生, 苏秀榕. 5个泥蚶群体遗传多样性的RAPD分析[J]. 生物多样性, 2003, 11(2): 118-124. |
| [13] | 高述民李凤兰陆帼一 杜慧芳赵英. 中国大蒜(Allium sativumL.)18个品种的酯酶同工酶多态性分析[J]. 植物学报, 2003, 20(06): 723-729. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||