

植物学报 ›› 2018, Vol. 53 ›› Issue (6): 782-792.DOI: 10.11983/CBB17258 cstr: 32102.14.CBB17258
罗俊杰1,2, 王莹1,2, 商辉1,3, 周喜乐4, 韦宏金1,3, 黄素楠2, 顾钰峰1,2, 金冬梅1,3, 戴锡玲2, 严岳鸿1,3,*( )
)
收稿日期:2017-12-29
出版日期:2018-11-01
发布日期:2018-12-05
通讯作者:
严岳鸿
作者简介:作者简介:白克智, 1959年开始在中国科学院植物研究所工作, 先后任助理研究员、研究员, 长期从事植物生长发育及其调控的研究。1986年,其主持的“满江红生物学特性研究”荣获中国科学院科技进步二等奖。曾任《植物生理学报》编委、《植物学报》常务编委、中国植物生长调节剂协会主任等职。
基金资助:
Luo Junjie1,2, Wang Ying1,2, Shang Hui1,3, Zhou Xile4, Wei Hongjin1,3, Huang Sunan2, Gu Yufeng1,2, Jin Dongmei1,3, Dai Xiling2, Yan Yuehong1,3,*( )
)
Received:2017-12-29
Online:2018-11-01
Published:2018-12-05
Contact:
Yan Yuehong
摘要: 孢粉学是解决植物分类中疑难类群物种微形态分化的重要方法, 随着分子系统学的发展, 结合这两门学科的优势可以更加有效地解决疑难类群的分类学问题。鳞盖蕨属(Microlepia)是一个分类困难的疑难类群, 采用孢粉学与分子系统学一一对应的方法, 以及居群取样方式, 选取280份样本, 联合4个叶绿体片段(rbcL、trnL-F、psbA-trnH和rps4), 采用最大似然法和贝叶斯法构建该属的系统发生关系, 在此基础上对凭证标本中100份材料的孢子进行观察和分析。综合分子系统学和孢粉学的研究结果, 得出结论: (1) 在形态学研究中广泛被接受的15个物种得到了单系支持, 并厘清了分类困难的复合群; (2) 发现边缘鳞盖蕨(M. marginata)可能存在隐性种; (3) 建议恢复过去归并处理为异名的瑶山鳞盖蕨(M. yaoshanica)、罗浮鳞盖蕨(M. lofoushanensis)、四川鳞盖蕨(M. szechuanica)以及滇西鳞盖蕨(M. subspeluncae); (4) 提出鳞盖蕨属可能存在杂交现象; (5) 提出鳞盖蕨属完整的属下分类建议。
罗俊杰, 王莹, 商辉, 周喜乐, 韦宏金, 黄素楠, 顾钰峰, 金冬梅, 戴锡玲, 严岳鸿. 基于孢子形态和分子证据探讨鳞盖蕨属(碗蕨科)系统分类. 植物学报, 2018, 53(6): 782-792.
Luo Junjie, Wang Ying, Shang Hui, Zhou Xile, Wei Hongjin, Huang Sunan, Gu Yufeng, Jin Dongmei, Dai Xiling, Yan Yuehong. Phylogeny and Systematics of the Genus Microlepia (Dennstaedtiaceae) based on Palynology and Molecular Evidence. Chinese Bulletin of Botany, 2018, 53(6): 782-792.
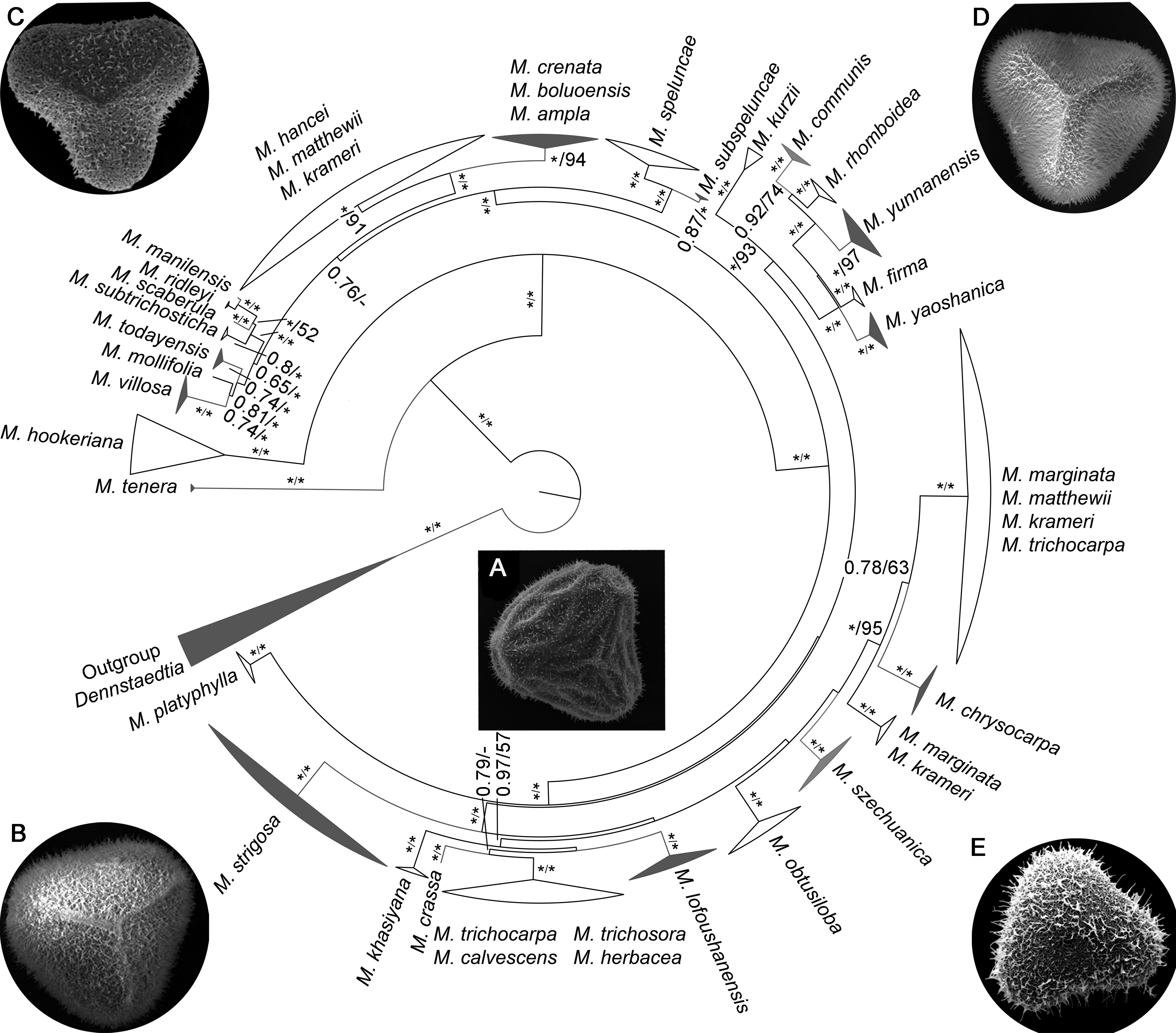
图 1 基于4个叶绿体片段rbcL、trnL-F、psbA-trnH和rps4联合数据构建的鳞盖蕨属贝叶斯系统发育树分支旁边的数字表示贝叶斯后验概率与ML自展支持率(≥50%); * 表示1或100; - 表示<0.5或50%。
Figure 1 Bayesian inference phylogeny of Microlepia derived from the combined data (rbcL, trnL-F, psbA-trnH and rps4) Values beside each branch represent bootstrap support for parsimony, Bayesian posterior probabilities and maximum likelihood (≥50%); * means 1 or 100; - means <0.5 or 50%.
| [1] | 曹建国, 于晶, 王全喜 (2007). 中国蕨类植物孢子的形态VII. 桫椤科. 云南植物研究 29, 7-12. |
| [2] | 陈焕镛 (1964). 海南植物志, 第1卷. 北京: 科学出版社. pp. 46-52. |
| [3] | 广西植物研究所 (2013). 广西植物志. 南宁: 广西科学技术出版社. pp. 96-108. |
| [4] | 孔宪需 (1988). 四川植物志, 第6卷. 蕨类植物门. 成都: 四川科学技术出版社. pp. 183-190. |
| [5] |
刘红梅, 张宪春, 曾辉 (2009). DNA序列在蕨类分子系统学研究中的应用. 植物学报 44, 143-158.
DOI URL |
| [6] | 牟善杰 (2010). 碗蕨科鳞盖蕨属专论研究. 博士论文. 台北: 台湾师范大学. pp. 1-198. |
| [7] | 秦仁昌 (1959). 中国植物志, 第2卷. 北京: 科学出版社. pp. 207-246. |
| [8] | 王培善, 王筱英 (2001). 贵州蕨类植物志. 贵阳: 贵州科技出版社. pp. 433-442. |
| [9] | 王全喜, 陈立群, 包文美 (1997). 中国金毛裸蕨属植物孢子形态的研究. 西北植物学报 17(5), 44-47. |
| [10] | 王全喜, 戴锡玲 (2010). 中国水龙骨目(真蕨目)植物孢子形态的研究. 北京: 科学出版社. pp. 1-25. |
| [11] |
王全喜, 于晶 (2003). 扫描电镜下真蕨目孢子表面纹饰的分类. 云南植物研究 25, 313-320.
DOI URL |
| [12] | 吴德邻 (2006). 广东植物志, 第7卷. 广州: 广东科技出版社. pp. 81-85. |
| [13] | 吴征镒 (2006). 云南植物志, 第12卷. 北京: 科学出版社. pp. 216-230. |
| [14] | 杨鲁红 (2012). 中国石韦属植物系统分类学研究. 硕士论文. 昆明: 云南大学. pp. 1-142. |
| [15] |
张宪春, 卫然, 刘红梅, 何丽娟, 王丽, 张钢民 (2013). 中国现代石松类和蕨类的系统发育与分类系统. 植物学报 48, 119-137.
DOI URL |
| [16] | 张玉龙, 席以珍, 张金谈, 高桂珍, 杜乃秋, 孙湘君, 孔昭宸 (1976). 中国蕨类植物孢子形态. 北京: 科学出版社. pp. 1-112. |
| [17] |
Akaike H (1974). A new look at the statistical model identification.IEEE Trans Automat Contr 19, 716-723.
DOI URL |
| [18] | Burland TG (2000). Dnastar’s lasergene sequence analysis software. In: Misener S, Krawetz SA, eds. Bioinformatics Methods and Protocols. Methods in Molecular BiologyTM, Vol. 132. Totowa, NJ: Humana Press. pp. 71-91. |
| [19] |
Denk T, Grimm GW (2009). Significance of pollen characteristics for infrageneric classification and phylogeny in Quercus(Fagaceae). Int J Plant Sci 170, 926-940.
DOI URL |
| [20] |
Ebihara A, Nitta JH, Ito M (2010). Molecular species identification with rich floristic sampling: DNA barcoding the pteridophyte flora of Japan.PLoS One 5, e15136.
DOI URL PMID |
| [21] | Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT.Nucl Acids Sym Ser 41, 95-98. |
| [22] | Hasebe M, Omori T, Nakazawa M, Sano T, Kato M, Iwatsuki K (1994). rbcL gene sequences provide evidence for the evolutionary lineages of leptosporangiate ferns. Proc Natl Acad Sci USA 91, 5730-5734. |
| [23] | Huang TC (1981). Spore Flora of Taiwan (Pteridophyta). Taiwan: National Taiwan University. pp. 52-53. |
| [24] | Kramer KU (1990). Dennstaedtiaceae. In: Kramer KU, Green PS, eds. The Families and Genera of Vascular Plants, Vol. I. Pteridophytes and Gymnosperms. Berlin: Springer-Verlag. pp. 81-94. |
| [25] |
Lehtonen S, Wahlberg N, Christenhusz MJM (2012). Diversification of lindsaeoid ferns and phylogenetic uncertainty of early polypod relationships.Bot J Linn Soc 170, 489-503.
DOI URL |
| [26] |
Li FW, Kuo LY, Rothfels CJ, Ebihara A, Chiou WL, Windham MD, Pryer KM (2011). rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS One 6, e26597.
DOI URL PMID |
| [27] | Miller MA, Pfeiffer W, Schwartz T (2010). Creating the cipres science gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE). New Orleans: IEEE. pp. 1-8. |
| [28] | Moran RC, Hanks JG, Rouhan G (2007). Spore morpho- logy in relation to phylogeny in the fern genus Elaphoglossum(Dryopteridaceae). Int J Plant Sci 168, 905-929. |
| [29] | Nakato N, Ebihara A (2011). Chromosome number of Microlepia hookeriana (Dennstaedtiaceae) and chromosome number evolution in the genus Microlepia. Bull Natl Mus Nat Sci Ser B 37, 75-78. |
| [30] |
Posada D (2008). Jmodeltest: phylogenetic model averaging.Mol Biol Evol 25, 1253-1256.
DOI URL |
| [31] |
Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R (2004). Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences.Am J Bot 91, 1582-1598.
DOI URL PMID |
| [32] |
Pryer KM, Smith AR, Hunt JS, Dubuisson JY (2001). rbcL data reveal two monophyletic groups of filmy ferns(Filicopsida: Hymenophyllaceae). Am J Bot 88, 1118-1130.
DOI URL PMID |
| [33] | Rambaut A, Drummond AJ (2007). Tracer v1.4. . |
| [34] |
Ronquist F, Huelsenbeck JP (2003). Mrbayes 3: Bayesian phylogenetic inference under mixed models.Bioinforma- tics 19, 1572-1574.
DOI URL |
| [35] |
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space.Syst Biol 61, 539-542.
DOI URL PMID |
| [36] | Schuettpelz E, Korall P, Pryer KM (2006). Plastid atpA data provide improved support for deep relationships among ferns. Taxon 55, 897-906. |
| [37] |
Schuettpelz E, Pryer KM (2007). Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes.Taxon 56, 1037-1050.
DOI URL |
| [38] |
Schwarz G (1978). Estimating the dimension of a model.Ann Statist 6, 461-464.
DOI URL |
| [39] |
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu WS, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005). The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis.Am J Bot 92, 142-166.
DOI URL |
| [40] |
Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H, Wolf PG (2006). A classification for extant ferns.Taxon 55, 705-731.
DOI URL |
| [41] | Souza-Chies TT, Bittar G, Nadot S, Carter L, Besin E, Lejeune B (1997). Phylogenetic analysis of Iridaceae with parsimony and distance methods using the plastid gene rps4. Plant Syst Evol 204, 109-123. |
| [42] | Swofford DL (2003). Paup*: Phylogenetic Analysis Using Parsimony, Version 4.0 b10. |
| [43] |
Taberlet P, Gielly L, Pautou G, Bouvet J (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA.Plant Mol Biol 17, 1105-1109.
DOI URL |
| [44] | Tagawa M (1952). Fern miscellany (6).J Jpn Bot 27, 213-218. |
| [45] |
Tate JA, Simpson BB (2003). Paraphyly of Tarasa(Malvaceae) and diverse origins of the polyploid species. Syst Bot 28, 723-737.
DOI URL |
| [46] |
The Pteridophyte Phylogeny Group (2016). A community-derived classification for extant lycophytes and ferns.J Syst Evol 54, 563-603.
DOI URL |
| [47] | Tryon AF, Lugardon B (1991). Spores of the Pteridophyta. Berlin: Springer-Verlag. pp. 1-279. |
| [48] |
Wang FG, Liu HM, He CM, Yang DM, Xing FW (2015). Taxonomic and evolutionary implications of spore ornamentation in Davalliaceae.J Syst Evol 53, 72-81.
DOI URL |
| [49] |
Wolf PG (1995). Phylogenetic analyses of rbcL and nuclear ribosomal RNA gene sequences in Dennstaedtiaceae. Am Fern J 85, 306-327.
DOI URL |
| [50] |
Wolf PG (1997). Evaluation of atpB nucleotide sequences for phylogenetic studies of ferns and other pteridophytes. Am J Bot 84, 1429-1440.
DOI URL PMID |
| [51] |
Xu Y, Hu CM, Hao G (2016). Pollen morphology of Androsace(Primulaceae) and its systematic implications. J Syst Evol 54, 48-64.
DOI URL |
| [52] | Yan YH, Qi XP, Zhang XC (2013). Dennstaedtiaceae. In: Wu ZY, Raven PH, Hong DY, eds. Flora of China, Vol. 2-3. Pteridophytes. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press. pp. 147-168. |
| [53] | Yanez A, Marquez GJ, Morbelli MA (2016). Palynological analysis of Dennstaedtiaceae taxa from the paranaense phytogeographic province that produce trilete spores II: Microlepia speluncae and Pteridium arachnoideum. An Acad Bras Ciênc 88, 877-890. |
| [54] |
Yuan Y, Fu L, Ma CY (2012). Microlepia boluoensis sp. nov.(Dennsteadtiaceae) from Guangdong, China. Nord J Bot 30, 168-173.
DOI URL |
| [55] |
Zhou XM, Zhang LB (2015). A classification of Selaginella(Selaginellaceae) based on molecular (chloroplast and nuclear), macromorphological, and spore features. Taxon 64, 1117-1140.
DOI URL |
| [1] | 熊高明, 申国珍, 赵常明, 徐文婷, 王杨, 谢宗强, 李家湘, 徐耀粘, 李跃林, 陈芳清. 中国檵木灌丛植被类型及群落特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [2] | 张坤, 伊丽晴, 刘占军, 张志杰, 王顼, 李明乐, 苗百岭, 张景慧, 李智勇, 董雷, 梁存柱. 毛刺锦鸡儿群落特征及分类[J]. 植物生态学报, 2025, 49(植被): 0-. |
| [3] | 杜燕 刘鑫 张瀚曰 马少伟 包维楷. 中国高山松群系的群落特征[J]. 植物生态学报, 2025, 49(植被): 1-0. |
| [4] | 刘碧颖, 陈子豪, 源思浩, 周婷. 植被志书研编中图表绘制的常见问题与技术参考[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [5] | 熊高明, 申国珍, 徐文婷, 谢宗强, 李跃林, 徐耀粘, 陈芳清, 李家湘. 中国低山丘陵热性常绿阔叶灌丛主要类型及群落特征[J]. 植物生态学报, 2025, 49(植被): 1-. |
| [6] | 韦丹丹, 杜燕, 包维楷, 胡斌, 张瀚曰, 王瀚婕, 唐圆圆, 黄龙, 郭昌安, 刘鑫. 冬麻豆群落的地理分布、特征和分类[J]. 植物生态学报, 2025, 49(濒危植物的保护与恢复): 1-. |
| [7] | 陈颖, 王迎雪, 邓清雅, 李培杨, 肖自新, 许艳蓉, 邓传远. 海坛岛典型灌丛群落数量分类与排序[J]. 植物生态学报, 2025, 49(4): 653-666. |
| [8] | 周璇, 张生芳, 刘宁, 鲁玉杰, 郑斯竹, 杨晓军, 路园园, 刘梅柯, 白明. 储藏物甲虫系统地位厘清及拉英汉名录更新[J]. 生物多样性, 2025, 33(4): 24238-. |
| [9] | 洪德元. 分类学中的方法论小叙[J]. 生物多样性, 2025, 33(2): 24541-. |
| [10] | 李邦泽, 张树仁. 中国莎草科最新物种名录和分类纲要[J]. 生物多样性, 2024, 32(7): 24106-. |
| [11] | 王腾, 李纯厚, 王广华, 赵金发, 石娟, 谢宏宇, 刘永, 刘玉. 西沙群岛七连屿珊瑚礁鱼类的物种组成与演替[J]. 生物多样性, 2024, 32(6): 23481-. |
| [12] | 吴琪, 张晓青, 杨雨婷, 周艺博, 马毅, 许大明, 斯幸峰, 王健. 浙江钱江源-百山祖国家公园庆元片区叶附生苔多样性及其时空变化[J]. 生物多样性, 2024, 32(4): 24010-. |
| [13] | 王淏, 钦鹏, 李仕贵. 水稻“混血杂交”群体揭示遗传互作奥秘[J]. 植物学报, 2024, 59(4): 529-532. |
| [14] | 董劭琼, 侯东杰, 曲孝云, 郭柯. 柴达木盆地植物群落样方数据集[J]. 植物生态学报, 2024, 48(4): 534-540. |
| [15] | 陈香蕾, 崔树娟, 赵晨军, 顾洪亮, 陈晓萍, 李锦隆, 孙俊. 基于叶形分类的木本植物单叶片面积预测模型[J]. 植物生态学报, 2024, 48(12): 1683-1691. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||