

植物学报 ›› 2018, Vol. 53 ›› Issue (3): 313-321.DOI: 10.11983/CBB17193 cstr: 32102.14.CBB17193
所属专题: 药用植物专辑 (2018年53卷3期)
任梦云1,2, 杜乐山1, 陈彦君1,2, 张盾2, 沈奇2, 关潇1,*( ), 张银东2,*(
), 张银东2,*( )
)
收稿日期:2017-10-18
接受日期:2018-04-30
出版日期:2018-05-01
发布日期:2018-09-11
通讯作者:
关潇,张银东
Ren Mengyun1,2, Du Leshan1, Chen Yanjun1,2, Zhang Dun2, Shen Qi2, Guan Xiao1,*( ), Zhang Yindong2,*(
), Zhang Yindong2,*( )
)
Received:2017-10-18
Accepted:2018-04-30
Online:2018-05-01
Published:2018-09-11
Contact:
Guan Xiao,Zhang Yindong
摘要: 为阐明锁阳(Cynomorium songaricum)的遗传结构与遗传多样性, 以甘肃省河西走廊地区及青海省共18个居群的188个锁阳个体为研究对象, 利用现代分子生物学技术, 采用序列分析方法从核-质基因方面对遗传结构进行了分析。结果显示, 锁阳ITS序列总长度为687 bp, 含有7个变异位点, 定义9个单倍型, 整体单倍型多态性Hd=0.294 20, 核苷酸多样性π=0.000 49。在整个单倍型网络中介图中, 单倍型H1位于中心位置, 并在所有的居群中均有分布, 为核心的古老单倍型。分子方差分析结果显示, 锁阳种群变异主要来源于种群内。根据ITS序列得到的群体间遗传分化系数以及Mantel检验结果, 锁阳种群间的遗传距离与地理距离之间不存在相关性, 表明现存的锁阳居群是相对近期发生生境片段化的产物。中性检验结果表明, 锁阳拒绝中性进化, 群体历经扩张或者基因座位受到负选择作用, 其中性零假说不能被排除。该研究为锁阳的系统分类、资源鉴定以及保护措施的制定提供了分子证据。
任梦云, 杜乐山, 陈彦君, 张盾, 沈奇, 关潇, 张银东. 锁阳ITS序列遗传多样性分析. 植物学报, 2018, 53(3): 313-321.
Ren Mengyun, Du Leshan, Chen Yanjun, Zhang Dun, Shen Qi, Guan Xiao, Zhang Yindong. Analysis on Genetic Diversity of Cynomorium songaricum by ITS Sequence. Chinese Bulletin of Botany, 2018, 53(3): 313-321.
| Sample No. | Location | Longitude E (°) | Latitude N (°) | Height (m) | Number | |
|---|---|---|---|---|---|---|
| R1 | Zhangye, Gansu | 102.522 | 36.822 | 1803 | 11 | |
| R2 | Minqin, Gansu | 100.772 | 39.223 | 1370 | 17 | |
| R3 | Jinchang, Gansu | 102.54 | 38.44 | 1466 | 7 | |
| R4 | Sandan, Gansu | 101.118 | 38.897 | 2020 | 10 | |
| R5 | Gaotai, Gansu | 99.086 | 39.787 | 1339 | 11 | |
| R6 | Sunan, Gansu | 99.176 | 39.604 | 1395 | 10 | |
| R7 | Sunan, Gansu | 99.474 | 39.445 | 1391 | 12 | |
| R8 | Jiuquan, Gansu | 98.55 | 40.3 | 1232 | 4 | |
| R9 | Yumen, Gansu | 97.196 | 40.516 | 1395 | 10 | |
| R10 | Guazhou, Gansu | 95.585 | 40.212 | 1343 | 11 | |
| R11 | Dunhuang, Gansu | 94.583 | 40.368 | 1033 | 11 | |
| R12 | Subei, Gansu | 96.619 | 39.397 | 2355 | 10 | |
| R13 | Delingha, Qinghai | 97.334 | 37.206 | 2780 | 11 | |
| R14 | Geermu, Qinghai | 92.839 | 36.701 | 2790 | 7 | |
| R15 | Dulan, Qinghai | 98.164 | 36.476 | 2972 | 12 | |
| R16 | Wulan, Qinghai | 98.622 | 36.458 | 2917 | 12 | |
| R17 | Gonghe, Qinghai | 100.183 | 36.446 | 2904 | 11 | |
| R18 | Guide, Qinghai | 101.639 | 36.219 | 2389 | 11 |
表1 18个锁阳居群样地分布信息
Table 1 Location of 18 populations of Cynomorium songaricum
| Sample No. | Location | Longitude E (°) | Latitude N (°) | Height (m) | Number | |
|---|---|---|---|---|---|---|
| R1 | Zhangye, Gansu | 102.522 | 36.822 | 1803 | 11 | |
| R2 | Minqin, Gansu | 100.772 | 39.223 | 1370 | 17 | |
| R3 | Jinchang, Gansu | 102.54 | 38.44 | 1466 | 7 | |
| R4 | Sandan, Gansu | 101.118 | 38.897 | 2020 | 10 | |
| R5 | Gaotai, Gansu | 99.086 | 39.787 | 1339 | 11 | |
| R6 | Sunan, Gansu | 99.176 | 39.604 | 1395 | 10 | |
| R7 | Sunan, Gansu | 99.474 | 39.445 | 1391 | 12 | |
| R8 | Jiuquan, Gansu | 98.55 | 40.3 | 1232 | 4 | |
| R9 | Yumen, Gansu | 97.196 | 40.516 | 1395 | 10 | |
| R10 | Guazhou, Gansu | 95.585 | 40.212 | 1343 | 11 | |
| R11 | Dunhuang, Gansu | 94.583 | 40.368 | 1033 | 11 | |
| R12 | Subei, Gansu | 96.619 | 39.397 | 2355 | 10 | |
| R13 | Delingha, Qinghai | 97.334 | 37.206 | 2780 | 11 | |
| R14 | Geermu, Qinghai | 92.839 | 36.701 | 2790 | 7 | |
| R15 | Dulan, Qinghai | 98.164 | 36.476 | 2972 | 12 | |
| R16 | Wulan, Qinghai | 98.622 | 36.458 | 2917 | 12 | |
| R17 | Gonghe, Qinghai | 100.183 | 36.446 | 2904 | 11 | |
| R18 | Guide, Qinghai | 101.639 | 36.219 | 2389 | 11 |
| Haplotype | Variable sites | Abundance | ||||||
|---|---|---|---|---|---|---|---|---|
| 63 | 86 | 197 | 274 | 421 | 605 | 623 | ||
| H1 | G | G | G | T | G | C | G | 143 |
| H2 | A | G | G | T | G | C | T | 3 |
| H3 | G | A | G | T | G | C | G | 1 |
| H4 | G | A | G | T | G | G | G | 2 |
| H5 | A | G | G | T | G | C | G | 26 |
| H6 | G | G | A | T | G | C | G | 5 |
| H7 | G | G | A | G | G | C | G | 1 |
| H8 | G | G | G | T | G | G | G | 4 |
| H9 | G | G | G | T | A | C | G | 3 |
表2 锁阳核糖体ITS序列的单倍型多态性位点
Table 2 Variable sites of ITS sequence haplotypes of Cynomorium songaricum
| Haplotype | Variable sites | Abundance | ||||||
|---|---|---|---|---|---|---|---|---|
| 63 | 86 | 197 | 274 | 421 | 605 | 623 | ||
| H1 | G | G | G | T | G | C | G | 143 |
| H2 | A | G | G | T | G | C | T | 3 |
| H3 | G | A | G | T | G | C | G | 1 |
| H4 | G | A | G | T | G | G | G | 2 |
| H5 | A | G | G | T | G | C | G | 26 |
| H6 | G | G | A | T | G | C | G | 5 |
| H7 | G | G | A | G | G | C | G | 1 |
| H8 | G | G | G | T | G | G | G | 4 |
| H9 | G | G | G | T | A | C | G | 3 |
| Population code | Samples | Haptotypes | Hd |
|---|---|---|---|
| R1 | 11 | H1, H2, H3, H4 | 0.25974 |
| R2 | 17 | H1, H5, H6 | 0.11586 |
| R3 | 7 | H1, H5 | 0.43956 |
| R4 | 10 | H1, H2, H5 | 0.42632 |
| R5 | 11 | H1, H6 | 0.24675 |
| R6 | 10 | H1, H7, H8 | 0.19474 |
| R7 | 12 | H1, H5 | 0.08333 |
| R8 | 4 | H1, H5, H9 | 0.60714 |
| R9 | 10 | H1, H8 | 0.10000 |
| R10 | 11 | H1, H5, H8 | 0.17749 |
| R11 | 11 | H1, H4, H6 | 0.17749 |
| R12 | 10 | H1 | 0 |
| R13 | 11 | H1 | 0 |
| R14 | 7 | H1, H6 | 0.14286 |
| R15 | 12 | H1, H9 | 0.08333 |
| R16 | 12 | H1, H9 | 0.08333 |
| R17 | 11 | H1, H8 | 0.09091 |
| R18 | 11 | H1 | 0 |
| Total | 188 | 0.29420 |
表3 锁阳18个居群的单倍型遗传多样性组成
Table 3 Haplotype diversity and composition of Cynomorium songaricum from18 populations
| Population code | Samples | Haptotypes | Hd |
|---|---|---|---|
| R1 | 11 | H1, H2, H3, H4 | 0.25974 |
| R2 | 17 | H1, H5, H6 | 0.11586 |
| R3 | 7 | H1, H5 | 0.43956 |
| R4 | 10 | H1, H2, H5 | 0.42632 |
| R5 | 11 | H1, H6 | 0.24675 |
| R6 | 10 | H1, H7, H8 | 0.19474 |
| R7 | 12 | H1, H5 | 0.08333 |
| R8 | 4 | H1, H5, H9 | 0.60714 |
| R9 | 10 | H1, H8 | 0.10000 |
| R10 | 11 | H1, H5, H8 | 0.17749 |
| R11 | 11 | H1, H4, H6 | 0.17749 |
| R12 | 10 | H1 | 0 |
| R13 | 11 | H1 | 0 |
| R14 | 7 | H1, H6 | 0.14286 |
| R15 | 12 | H1, H9 | 0.08333 |
| R16 | 12 | H1, H9 | 0.08333 |
| R17 | 11 | H1, H8 | 0.09091 |
| R18 | 11 | H1 | 0 |
| Total | 188 | 0.29420 |
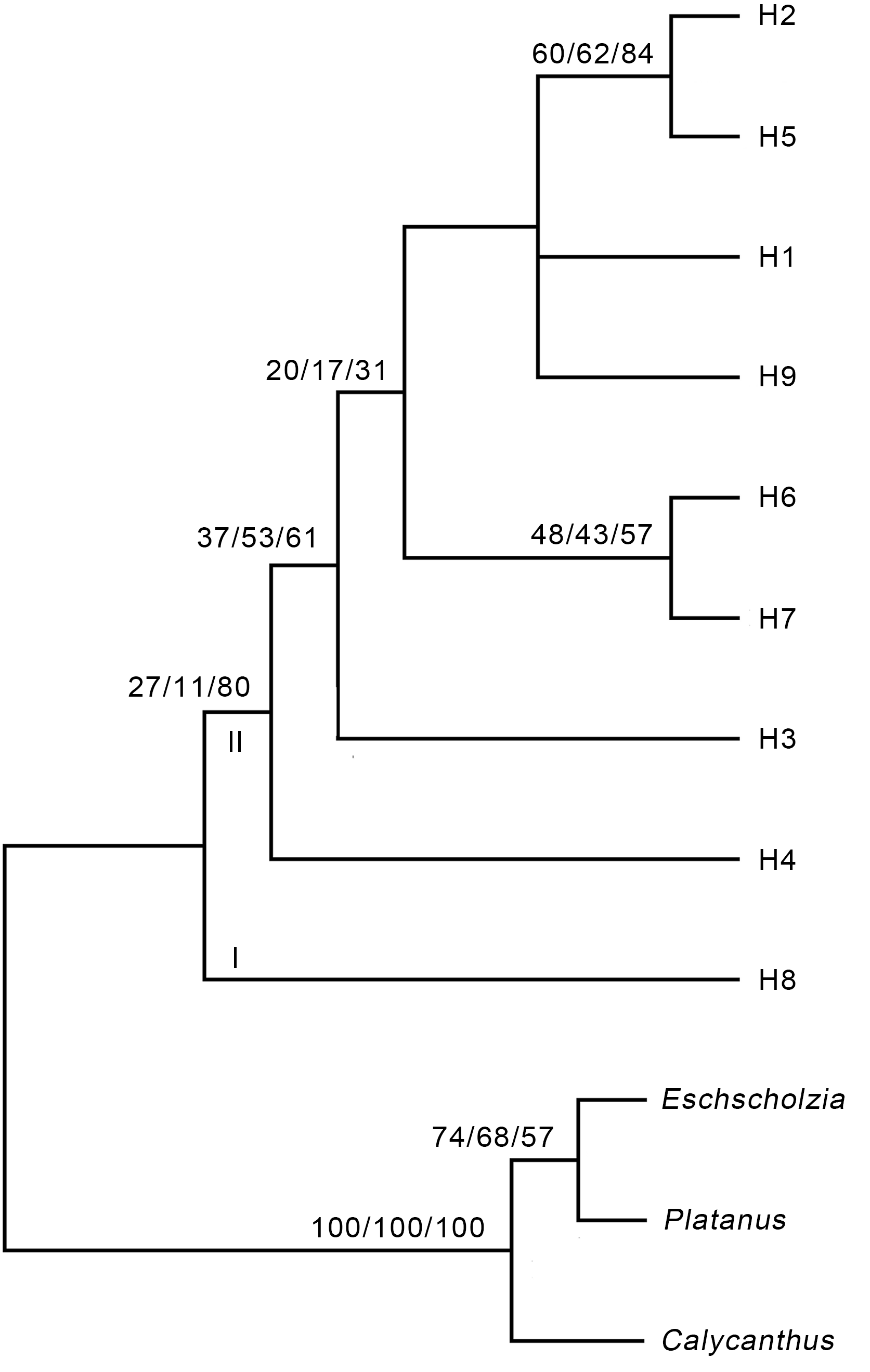
图1 基于锁阳ITS序列的严格一致性树步长=784, CI=0.949 0, RI=0.911 7, RCI=0.865 2。分支上的数字分别代表MP/ML/BI的支持率。
Figure 1 Strict consensus tree based on the ITS sequence of Cynomorium songaricumLength=784, CI=0.949 0, RI=0.911 7, RCI=0.865 2. The num- bers on the branch represent the support rate of MP/ML/BI, respectively.
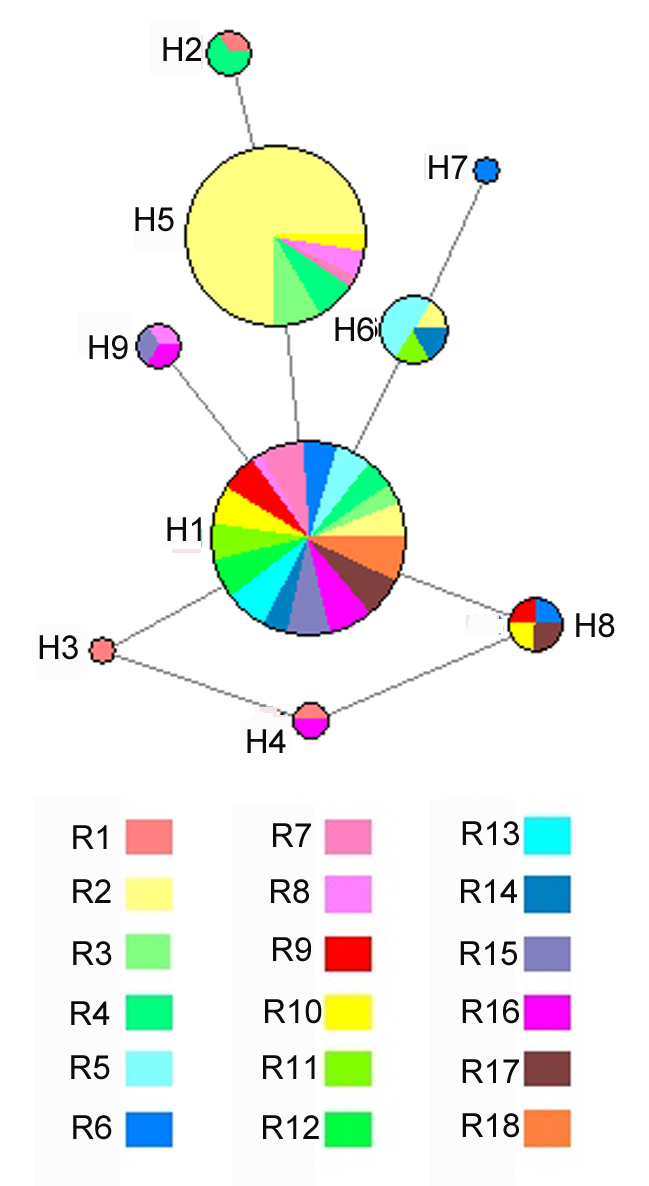
图2 锁阳18个居群基于ITS序列的单倍型网络图圆圈的大小与单倍型的相对频率成正比; 不同颜色代表锁阳属不同种群。
Figure 2 Haplotype network based on ITS sequence of Cynomorium songaricum from 18 populationsThe size of circles are proportional to the relative frequency of the haplotype; Different colors represent different populations of Cynomorium songaricum.
| Sequence | HS (SE) | HT (SE) | GST (SE) | NST (SE) |
|---|---|---|---|---|
| ITS | 0.179 (0.0390) | 0.277 (0.0820) | 0.353 (0.1914) | 0.408 (0.2038) |
表4 锁阳种群遗传结构参数
Table 4 Genetic structural parameters of Cynomorium songaricum
| Sequence | HS (SE) | HT (SE) | GST (SE) | NST (SE) |
|---|---|---|---|---|
| ITS | 0.179 (0.0390) | 0.277 (0.0820) | 0.353 (0.1914) | 0.408 (0.2038) |
| Source of variation | df | Sum of squares | Variance components | Percentage | FST | P |
|---|---|---|---|---|---|---|
| Among populations | 17 | 28.833 | 0.07689 Va | 44.57 | 0.44566 | <0.001* |
| Within populations | 358 | 34.239 | 0.09564 Vb | 55.43 | ||
| Total | 375 | 63.072 | 0.17253 |
表5 锁阳核糖体基因单倍型分子方差分析
Table 5 Analysis of molecular variance for ribosome haplotypes of Cynomorium songaricum
| Source of variation | df | Sum of squares | Variance components | Percentage | FST | P |
|---|---|---|---|---|---|---|
| Among populations | 17 | 28.833 | 0.07689 Va | 44.57 | 0.44566 | <0.001* |
| Within populations | 358 | 34.239 | 0.09564 Vb | 55.43 | ||
| Total | 375 | 63.072 | 0.17253 |
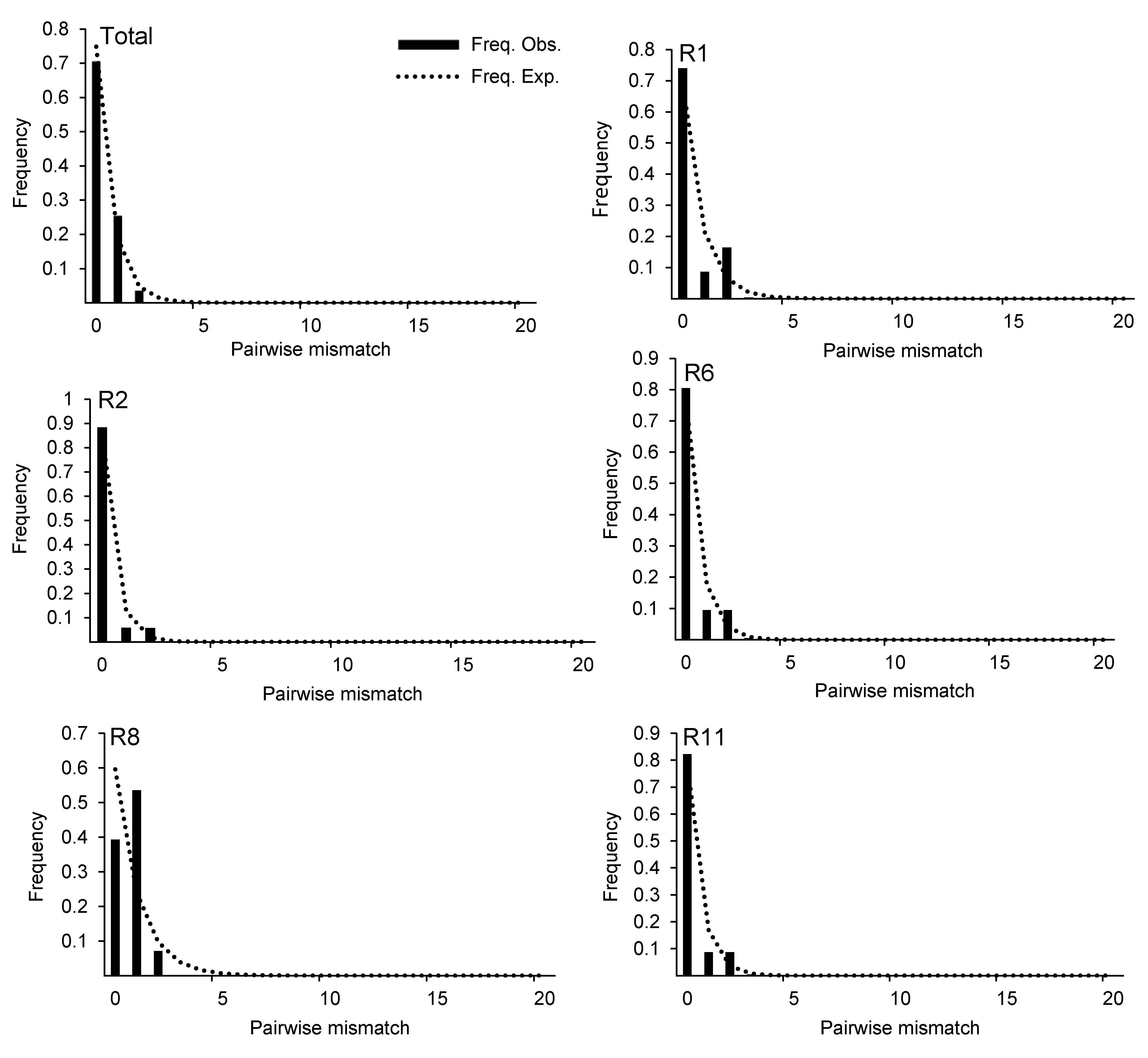
图3 基于ITS序列的锁阳种群失配分析柱形代表变异位点在群体扩张模型下的预期分布; 虚线代表变异位点的实际分布。
Figure 3 Mismatch distribution analysis for the populations of Cynomorium songaricum based on ITS sequenceCylindricality represents the expected distribution of variation sites under the population expansion model; dotted line represents the actual distribution of variation sites.
| [1] | 陈贵林, 岳鑫, 刘广达 (2011). 锁阳愈伤组织体系的建立及遗传多样性研究. 见: 第十届全国药用植物及植物药学术研讨会论文集. 昆明: 中国植物学会. pp. 26. |
| [2] | 陈叶, 高海宁, 高宏, 韩多宏, 罗光宏, 张勇 (2013). 甘肃河西走廊道地药材锁阳的分布和利用. 中兽医医药杂志 32, 77-79. |
| [3] | 郝媛媛, 岳利军, 康建军, 王锁民 (2012). “沙漠人参”肉苁蓉和锁阳研究进展. 草业学报21, 286-293. |
| [4] | 黄建峰, 李朗, 李捷 (2016). 樟属植物ITS序列多态性分析. 植物学报 51, 609-619. |
| [5] | 李洪芹, 马昌豪, 彭艳丽 (2015). 山东玫瑰花核糖体rDNA ITS序列分析初步探究. 天津中医药大学学报 34, 104-107. |
| [6] | 李金博, 高丽华, 周美亮, 李诗刚, 彭昭良, 吴燕民 (2016). 15个白三叶品种的ISSR和ITS遗传多样性分析. 草业科学 33, 1147-1153. |
| [7] | 李学营, 彭建营, 白瑞霞 (2005). 基于核rDNA的ITS序列在种子植物系统发育研究中的应用. 西北植物学报 25, 829-834. |
| [8] | 宁淑萍, 颜海飞, 郝刚, 葛学军 (2008). 植物DNA条形码研究进展. 生物多样性 16, 417-425. |
| [9] | 宋宇婷, 马大龙, 隋心, 穆立蔷 (2011). 黑龙江、吉林地区松茸ITS序列遗传多样性分析. 中国酿造 30, 133-136. |
| [10] | 岳鑫, 段园园, 陈贵林 (2013). 锁阳愈伤组织诱导和增殖及不定根分化. 植物生理学报 49, 1421-1426. |
| [11] | 中国科学院中国植物志编辑委员会 (2000). 中国植物志(第五十三卷第二分册). 北京: 科学出版社. pp. 152-154. |
| [12] | Avise JC (2000).Phylogeography: the History and Formation of Species. Cambridge: Harvard University Press. pp. 134-135. |
| [13] | Bandelt HJ, Forster P, Röhl A (1999). Median-joining networks for inferring intraspecific phylogenies.Mol Biol Evol 16, 37-48. |
| [14] | Barkman TJ, McNeal JR, Lim SH, Coat G, Croom HB, Young ND, Depamphilis CW (2007). Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants.BMC Evol Biol 7, 248. |
| [15] | Cibrián-Jaramillo A, Bacon CD, Garwood NC, Bateman RM, Thomas MM, Russell S, Bailey CD, Hahn WJ, Bridgewater SG, DeSalle R (2009). Population genetics of the understory fishtail palm Chamaedorea ernestiau- gusti in Belize: high genetic connectivity with local differen- tiation. BMC Genet 10, 65. |
| [16] | Cooke DEL, Duncan JM (1997). Phylogenetic analysis of Phytophthora species based on ITS1 and ITS2 sequences of the ribosomal RNA gene repeat. Mycol Res 101, 667-677. |
| [17] | Cosacov A, Johnson LA, Paiaro V, Cocucci AA, Córdoba FE, Sérsic AN, Crisci J (2013). Precipitation rather than temperature influenced the phylogeography of the endemic shrub Anarthrophyllum desideratum in the Patagonian steppe. J Biogeogra 40, 168-182. |
| [18] | Excoffier L (2004). Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model.Mol Ecol 13, 853-864. |
| [19] | Excoffier L, Laval G, Schneider S (2007). Arlequin (version 3.0): an integrated software package for population genetics data analysis.Evol Bioinform 1, 47-50. |
| [20] | Excoffier L, Smouse PE, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data.Genetics 131, 479-491. |
| [21] | Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/ 98/NT. In: Information Retrieval Ltd., Nucleic Acids Symposium Series, vol. 41. London: IRL Press. pp. 95-98. |
| [22] | Hou DY, Song JY, Shi LC, Ma XC, Xin TY, Han JP, Xiao W, Sun ZY, Cheng RY, Yao H (2013a). Stability and accuracy assessment of identification of traditional Chinese materia medica using DNA barcoding: a case study on Flos Lonicerae Japonicae.Biomed Res Int 2013, 549037. |
| [23] | Hou DY, Song JY, Yao H, Han JP, Pang XH, Shi LC, Wang XC, Chen SL (2013b). Molecular identification of corni fructus and its adulterants by ITS/ITS2 sequences.Chin J Nat Med 11, 121-127. |
| [24] | Kumar S, Stecher G, Tamura K (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger data- sets.Mol Biol Evol 33, 1870-1874. |
| [25] | Lahaye R, Van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008). DNA barcoding the floras of biodiversity hotspots.Proc Natl Acad Sci USA 105, 2923-2928. |
| [26] | Librado P, Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data.Bioinformatics 25, 1451-1452. |
| [27] | Liu GD, Chen GL, Li W, Li CX (2013). Genetic and phytochemical diversities of Cynomorium songaricum Rupr. in Northwest China indicated by ISSR markers and HPLC- fingerprinting. Biochem Syst Ecol 48, 34-41. |
| [28] | Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J (2008). Testing candidate plant barcode regions in the My- risticaceae.Mol Ecol Res 8, 480-490. |
| [29] | Nickrent DL, Der JP, Anderson FE (2005). Discovery of the photosynthetic relatives of the “Maltese mushroom” Cynomorium. BMC Evol Biol 5, 38. |
| [30] | Pons O, Petit RJ (1996). Measwring and testing genetic differentiation with ordered Versus unordered alleles. Genetics 144, 1237-1245. |
| [31] | Rogers AR, Harpending H (1992). Population growth makes waves in the distribution of pairwise genetic differences.Mol Biol Evol 9, 552-569. |
| [32] | Slatkin M, Hudson RR (1991). Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations.Genetics 129, 555-562. |
| [33] | Song JY, Shi LC, Li DZ, Sun YZ, Niu YY, Chen ZD, Luo HM, Pang XH, Sun ZY, Liu C, Lv AP, Deng YP, Larson-Rabin Z, Wilkinson M, Chen SL (2012). Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA.PLoS One 7, e43971. |
| [34] | Swafford DL (2002). PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4.0 10b. Sunderland, MA:Sinauer Associates. |
| [35] | Zhang ZH, Li CQ, Li JH (2009). Phylogenetic placement of Cynomorium in Rosales inferred from sequences of the inverted repeat region of the chloroplast genome. J Syst Evol 47, 297-304. |
| [1] | 黄承玲, 黎荣瀚, 覃红玲, 杨胜雄, 田晓玲, 夏国威, 陈正仁, 周玮. 基于SNP分子标记的极小种群野生植物荔波杜鹃保护遗传学研究[J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | 张如礼, 李德铢, 张玉霄. 短穗竹居群遗传结构及气候适应性分析[J]. 植物学报, 2025, 60(3): 407-424. |
| [3] | 杨志刚, 张鹏程, 常海文, 康立茹, 左毅, 向浩鑫, 韩凤英. 基于形态学性状和SSR标记的辣椒种质资源遗传多样性分析(长英文摘要)[J]. 植物学报, 2025, 60(2): 218-234. |
| [4] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [5] | 陈静, 张丙昌, 刘燕晋, 武杰, 赵康, 明姣. 荒漠生物结皮细鞘丝藻类(Leptolyngbya-like)蓝藻多样性[J]. 生物多样性, 2024, 32(9): 24186-. |
| [6] | 马佳正, 陈雨婷, 马松梅, 张丹, 贺凌云. 基于多源数据的新疆沙漠植物白梭梭遗传格局与扩散路径模拟[J]. 植物生态学报, 2024, 48(10): 1326-1335. |
| [7] | 李庆多, 栗冬梅. 全球蝙蝠巴尔通体流行状况分析[J]. 生物多样性, 2023, 31(9): 23166-. |
| [8] | 冯晨, 张洁, 黄宏文. 统筹植物就地保护与迁地保护的解决方案: 植物并地保护(parallel situ conservation)[J]. 生物多样性, 2023, 31(9): 23184-. |
| [9] | 齐海玲, 樊鹏振, 王跃华, 刘杰. 中国北方六省区胡桃的遗传多样性和群体结构[J]. 生物多样性, 2023, 31(8): 23120-. |
| [10] | 熊飞, 刘红艳, 翟东东, 段辛斌, 田辉伍, 陈大庆. 基于基因组重测序的长江上游瓦氏黄颡鱼群体遗传结构[J]. 生物多样性, 2023, 31(4): 22391-. |
| [11] | 蒲佳佳, 杨平俊, 戴洋, 陶可欣, 高磊, 杜予州, 曹俊, 俞晓平, 杨倩倩. 长江下游外来生物福寿螺的种类及其种群遗传结构[J]. 生物多样性, 2023, 31(3): 22346-. |
| [12] | 何艺玥, 刘玉莹, 张富斌, 秦强, 曾燏, 吕振宇, 杨坤. 梯级水利工程背景下的嘉陵江干流蛇鮈群体遗传多样性和遗传结构[J]. 生物多样性, 2023, 31(11): 23160-. |
| [13] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [14] | 陶克涛, 白东义, 图格琴, 赵若阳, 安塔娜, 铁木齐尔·阿尔腾齐米克, 宝音德力格尔, 哈斯, 芒来, 韩海格. 基于基因组SNPs对东亚家马不同群体遗传多样性的评估[J]. 生物多样性, 2022, 30(5): 21031-. |
| [15] | 陈天翌, 娄安如. 青藏高原东侧白桦种群的遗传多样性与遗传结构[J]. 植物生态学报, 2022, 46(5): 561-568. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||