

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (2): 175-184.DOI: 10.11983/CBB17144 cstr: 32102.14.CBB17144
• INVITED REVIEWS • Previous Articles Next Articles
Han Danlu, Lai Jianbin, Yang Chengwei*( )
)
Received:2017-08-07
Accepted:2017-10-20
Online:2018-03-01
Published:2018-03-10
Contact:
Yang Chengwei
Han Danlu, Lai Jianbin, Yang Chengwei. Research Advances in Functions of SUMO E3 Ligases in Plant Growth and Development[J]. Chinese Bulletin of Botany, 2018, 53(2): 175-184.
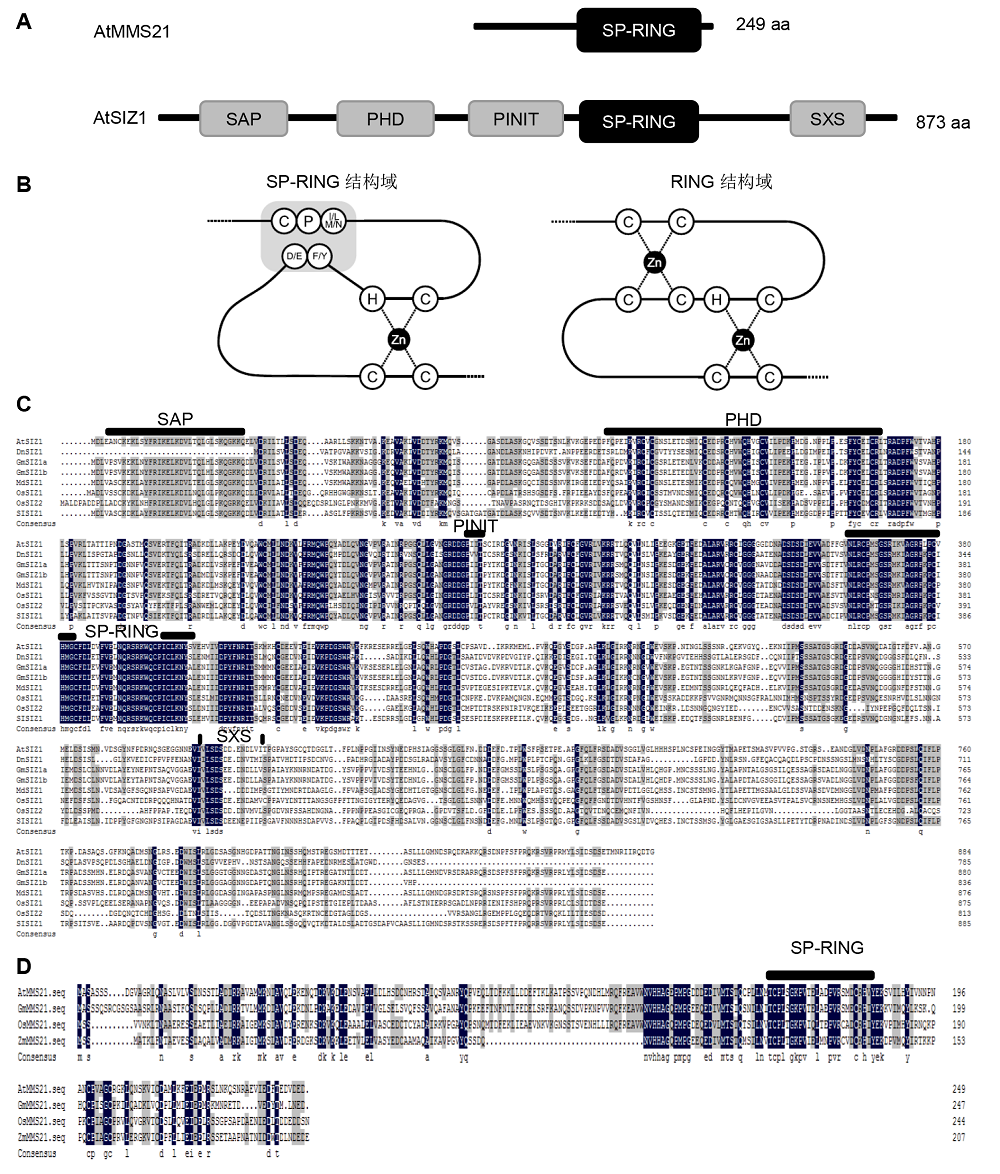
Figure 1 The domains are conserved in plant SUMO E3 ligases(A) Characteristic domains of plant SUMO E3 ligases. MMS21 and SIZ1 are shown by boxes, SIZ1 possesses SAP, PHD, PINIT, SP-RING and SXS domains, while MMS21 possesses only SP-RING domain (Ishida et al., 2012); (B) A schematic model of the SP-RING domain of SUMO E3 ligases and the RING finger of ubiquitin E3 ligases. The RING domain sports two zinc-coordinating loops, the SP-RING domain contains only one, the second loop is instead held together by hydrogen bonds and Van der Waals forces (Duan et al., 2009; Yunus and Lima, 2009; Ishida et al., 2012); (C), (D) The sequence data for the plant SIZ1 homologues were obtained from the NCBI protein database. Sequence identities (black boxes) and similarities (gray boxes) of amino acids were identified by DNAMAN. Amino acid sequences of SIZ1 proteins are from Arabidopsis thaliana AtSIZ1 (AAU00414.1), Glycine max GmSIZ1a (KRH24918.1), GmSIZ1b (KRG89029.1), Oryza sativa OsSIZ1 (BAG97182.1), OsSIZ2 (BAG89374.1), and Zea mays ZmSIZ1a (AQK95338.1), ZmSIZ1b (AQK79048.1), ZmSIZc (ONM41936.1). Amino acid sequences of MMS21 proteins are from Arabidopsis thaliana AtMMS21 (NP_188133.2), Glycine max GmMMS21 (XP_003541835.1), Oryza sativa OsMMS21 (XP_015640264.1), and Zea mays ZmMMS21 (AQK88509.1).
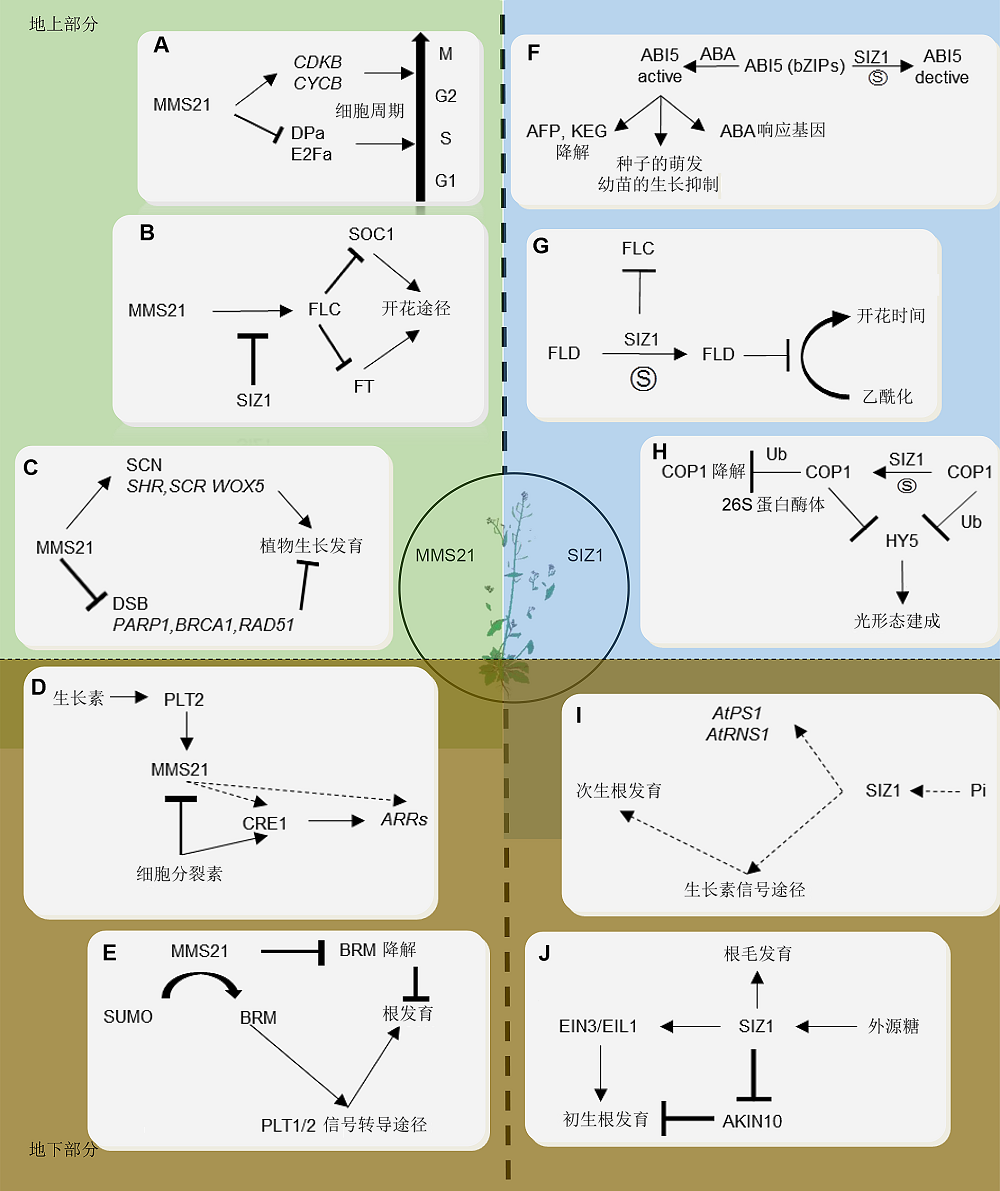
Figure 2 The functions of SUMO E3 ligases in Arabidopsis(A) The function of MMS21 regulates in cell cycle (Liu et al., 2016); (B) MMS21 and SIZ1 are both involved in flowering pathway and time (Kwak et al., 2016); (C) MMS21 responses to the DNA damage to regulate plant growth and development (Xu et al., 2013; Yuan et al., 2014); (D) MMS21 is involved in Arabidopsis root development by auxin (Ishida et al., 2009; Huang et al., 2009; Zhang et al., 2010); (E) MMS21 is involved in root development by regulating chromatin remodeling complex (Zhang et al., 2017a); (F) SIZ1 regulate glucose metabolism germinated/seedling development (Miura et al., 2009); (G) SIZ1 is involved in flowering pathway (Jin et al., 2008); (H) SIZ1 regulates the photomorphogenesis (Kim et al., 2016a; Lin et al., 2016); (I) SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation (Miura et al., 2005, 2011); (J) SIZ1 regulates development of root by sugar pathway (Castro et al., 2015)
| [1] | 徐庞连, 曾棉炜, 黄丽霞, 阳成伟 (2008). 植物SUMO化修饰及其生物学功能. 植物学通报 25, 608-615. |
| [2] | 杨辉霞, 童依平, 王道文 (2007). 拟南芥低磷胁迫反应分子机理研究的最新进展. 植物学通报 24, 726-734. |
| [3] | 石田喬志, 杉本慶子 (2012). 植物におけるSUMOシステムとE3リガーゼの機能. 生化学 84, 440-447. |
| [4] | Almedawar S, Colomina N, Bermúdez-lópez M, Pociño- Merino I, Torres-Rosell J (2012). A SUMO-dependent step during establishment of sister chromatid cohesion.Curr Biol 22, 1576-1581. |
| [5] | Cai B, Kong XX, Zhong C, Sun S, Zhou XF, Jin YH, Wang YN, Li X, Zhu ZD, Jin JB (2017). SUMO E3 ligases GmSIZ1a and GmSIZ1b regulate vegetative growth in soybean.J Integr Plant Biol 59, 2-14. |
| [6] | Cappadocia L, Pichler A, Lima CD (2015). Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase.Nat Struct Mol Biol 22, 968-975. |
| [7] | Castro PH, Verde N, Lourenço T, Magalhães AP, Tavares RM, Bejarano ER, Azevedo H (2015). SIZ1-dependent post-translational modification by SUMO modulates sugar signaling and metabolism in Arabidopsis thaliana. Plant Cell Physiol 56, 2297-2311. |
| [8] | Catala R, Ouyang J, Abreu IA, Hu YX, Seo H, Zhang XR, Chua NH (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses.Plant Cell 19, 2952-2966. |
| [9] | Cheong MS, Park HC, Hong MJ, Lee J, Choi W, Jin JB, Bohnert HJ, Lee SY, Bressan RA, Yun DJ (2009). Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes.Plant Phy- siol 151, 1930-1942. |
| [10] | De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, Farmer S, Hwang JY, Machin F, Ceschia A, McAleenan A, Cordon-Preciado V, Clemente-Blanco A, Vilella-Mitjana F, Ullal P, Jarmuz A, Leitao B, Bressan D, Dotiwala F, Papusha A, Zhao XL, Myung K, Haber JE, Aguilera A, Aragón L (2006). Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination.Nat Cell Biol 8, 1032-1034. |
| [11] | Desterro JMP, Rodriguez MS, Kemp GD, Hay RT (1999). Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1.J Biol Chem 274, 10618-10624. |
| [12] | Desterro JMP, Thomson J, Hay RT (1997). Ubch9 conjugates SUMO but not ubiquitin.FEBS Lett 417, 297-300. |
| [13] | Dong JS, Piñeros MA, Li XX, Yang HB, Liu Y, Murphy AS, Kochian LV, Liu D (2017). An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots.Mol Plant 10, 244-259. |
| [14] | Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H (2009). Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex.Mol Cell 35, 657-668. |
| [15] | Eisenhardt N, Chaugule VK, Koidl S, Droescher M, Dogan E, Rettich J, Sutinen P, Imanishi SY, Hofmann K, Palvimo JJ, Pichler A (2015). A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly.Nat Struct Mol Biol 22, 959-967. |
| [16] | Gong LM, Li B, Millas S, Yeh ETH (1999). Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex.FEBS Lett 448, 185-189. |
| [17] | Huang LX, Yang SG, Zhang SC, Liu M, Lai JB, Qi YL, Shi SF, Wang JX, Wang YQ, Xie Q, Yang CW (2009). The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root.Plant J 60, 666-678. |
| [18] | Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009). SUMO E3 ligase HIGH PLOIDY2 regul- ates endocycle onset and meristem maintenance in Arabidopsis.Plant Cell 21, 2284-2297. |
| [19] | Ishida T, Yoshimura M, Miura K, Sugimoto K (2012). MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development.PLoS One 7, e46897. |
| [20] | Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects onFLC chromatin structure. Plant J 53, 530-540. |
| [21] | Johnson ES, Blobel G (1997). Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p.J Biol Chem 272, 26799-26802. |
| [22] | Johnson ES, Gupta AA (2001). An E3-like factor that promotes SUMO conjugation to the yeast septins.Cell 106, 735-744. |
| [23] | Kim JY, Jang IC, Seo HS (2016a). COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity.Front Plant Sci 7, 1182. |
| [24] | Kim SI, Kwak JS, Song JT, Seo HS (2016b). The E3 SUMO ligase AtSIZ1 functions in seed germination in Arabidopsis.Physiol Plant 158, 256-271. |
| [25] | Kim SI, Park BS, Kim DY, Yeu SY, Song SI, Song JT, Seo HS (2015). E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signaling and plant development.Bio- chem J 469, 299-314. |
| [26] | Kwak JS, Son GH, Kim SI, Song JT, Seo HS (2016). Arabidopsis HIGH PLOIDY2 sumoylates and stabilizes Flowering Locus C through its E3 ligase activity.Front Plant Sci 7, 530. |
| [27] | Lin XL, Niu D, Hu ZL, Kim DH, Jin YH, Cai B, Liu P, Miura K, Yun DJ, Kim WY, Lin RC, Jin JB (2016). An Arabidopsis SUMO E3 ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity.PLoS Ge- net 12, e1006016. |
| [28] | Ling Y, Zhang CY, Chen T, Hao HQ, Liu P, Bressan RA, Hasegawa PM, Jin JB, Lin JX (2012). Mutation in SUMO E3 ligase, SIZ1, disrupts the mature female gametophyte in Arabidopsis.PLoS One 7, e29470. |
| [29] | Liu F, Wang X, Su MY, Yu MY, Zhang SC, Lai JB, Yang CW, Wang YQ (2015). Functional characterization of DnSIZ1, a SIZ/PIAS-type SUMO E3 ligase fromDendrobium. BMC Plant Biol 15, 225. |
| [30] | Liu M, Shi SF, Zhang SC, Xu PL, Lai JB, Liu YY, Yuan DK, Wang YQ, Du JJ, Yang CW (2014). SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis.BMC Plant Biol 14, 153. |
| [31] | Liu YY, Lai JB, Yu MY, Wang FG, Zhang JJ, Jiang JM, Hu H, Wu Q, Lu GH, Xu PL, Yang CW (2016). The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/ DPa complex in cell cycle regulation.Plant Cell 28, 2225-2237. |
| [32] | Miura K, Jin JB, Hasegawa PM (2007).a Sumoylation, a post-translational regulatory process in plants.Curr Opin Plant Biol 10, 495-502. |
| [33] | Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ash- worth EN, Bressan RA, Yun DJ, Hasegawa PM (2007).b SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Ara- bidopsis. Plant Cell 19, 1403-1414. |
| [34] | Miura K, Lee J, Gong QQ, Ma SS, Jin JB, Yoo CY, Miura T, Sato A, Bohnert HJ, Hasegawa PM (2011). SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiol 155, 1000-1012. |
| [35] | Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling.Proc Natl Acad Sci USA 106, 5418-5423. |
| [36] | Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses.Proc Natl Acad Sci USA 102, 7760-7765. |
| [37] | Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H (1999). In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun 254, 693-698. |
| [38] | Park HC, Kim H, Koo SC, Park HJ, Cheong MS, Hong H, Baek D, Chung WS, Kim DH, Bressan RA, Lee SY, Bohnert HJ, Yun DJ (2010). Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice.Plant Cell Environ 33, 1923-1934. |
| [39] | Pichler A, Fatouros C, Lee H, Eisenhardt N (2017). SUMO conjugation—a mechanistic view.Biomol Concepts 8, 13-36. |
| [40] | Reverter D, Lima CD (2005). Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex.Nature 435, 687-692. |
| [41] | Streich Jr FC, Lima CD (2016). Capturing a substrate in an activated RING E3/E2-SUMO complex.Nature 536, 304-308. |
| [42] | Thangasamy S, Guo CL, Chuang MH, Lai MH, Chen J, Jauh GY (2011). Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence.New Phytol 189, 869-882. |
| [43] | Wang HD, Sun R, Cao Y, Pei WX, Sun YF, Zhou HM, Wu XN, Zhang F, Luo L, Shen QR, Xu GH (2015). OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice. Plant Cell Physiol 56, 2381-2395 |
| [44] | Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I (2009). The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21, 2688-2699. |
| [45] | Wotton D, Pemberton LF, Merrill-Schools J (2017). SUMO and Chromatin Remodeling. In: Wilson VG, ed. SUMO Regulation of Cellular Processes. Cham: Springer. pp. 35-50. |
| [46] | Xu PL, Yuan DK, Liu M, Li CX, Liu YY, Zhang SC, Yao N, Yang CW (2013). AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots.Plant Physiol 161, 1755-1768. |
| [47] | Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM (2006). SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid.Plant Physiol 142, 1548-1558. |
| [48] | Yuan DK, Lai JB, Xu PL, Zhang SC, Zhang JJ, Li CX, Wang YQ, Du JJ, Liu YY, Yang CW (2014). AtMMS21 regulates DNA damage response and homologous recombination repair in Arabidopsis.DNA Repair 21, 140-147. |
| [49] | Yunus AA, Lima CD (2009). Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA.Mol Cell 35, 669-682. |
| [50] | Zhang JJ, Lai JB, Wang FG, Yang SG, He ZP, Jiang JM, Li QL, Wu Q, Liu YY, Yu MY, Du MY, Du JJ, Xie Q, Wu KQ (2017).a A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root deve- lopment.Plant Physiol 173 1574-1582. |
| [51] | Zhang RF, Guo Y, Li YY, Zhou LJ, Hao YJ, You CX (2016). Functional identification of MdSIZ1 as a SUMO E3 ligase in apple.J Plant Physiol 198, 69-80. |
| [52] | Zhang S, Zhuang KY, Wang SJ, Lv JL, Ma NN, Meng QW (2017).b A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco.J Integr Plant Biol 59, 102-117. |
| [53] | Zhang SC, Qi YL, Yang CW (2010). Arabidopsis SUMO E3 ligase AtMMS21 regulates root meristem development.Plant Signal Behav 5, 53-55. |
| [54] | Zheng Y, Schumaker KS, Guo Y (2012). Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109, 12822-12827. |
| [1] | Yaping Wang, Wenquan Bao, Yu’e Bai. Advances in the Application of Single-cell Transcriptomics in Plant Growth, Development and Stress Response [J]. Chinese Bulletin of Botany, 2025, 60(1): 101-113. |
| [2] | Xinhai Zeng, Rui Chen, Yu Shi, Chaoyue Gai, Kai Fan, Zhaowei Li. Research Advances in Biological Functions of Plant SPL Transcription Factors [J]. Chinese Bulletin of Botany, 2023, 58(6): 982-997. |
| [3] | Yanan Xu, Jiarong Yan, Xin Sun, Xiaomei Wang, Yufeng Liu, Zhouping Sun, Mingfang Qi, Tianlai Li, Feng Wang. Red and Far-red Light Regulation of Plant Growth, Development, and Abiotic Stress Responses [J]. Chinese Bulletin of Botany, 2023, 58(4): 622-637. |
| [4] | Huang Junwen, Feng Qiyi, Zheng Kaiyong, Huang Junjie, Wang Linbo, Lai Jianbin Lai Ruiqiang, Yang Chengwei. An Effective in Vitro SUMOylation Detection System for Plant Proteins [J]. Chinese Bulletin of Botany, 2022, 57(4): 490-499. |
| [5] | Jindong Wang,Yu Zhou,Jiawen Yu,Xiaolei Fan,Changquan Zhang,Qianfeng Li,Qiaoquan Liu. Advances in the Regulation of Plant Growth and Development and Stress Response by miR172-AP2 Module [J]. Chinese Bulletin of Botany, 2020, 55(2): 205-215. |
| [6] | Gaoping Qu,Jingbo Jin. Detection of SUMOylation in Plants [J]. Chinese Bulletin of Botany, 2020, 55(1): 83-89. |
| [7] | Menglong Wang,Xiaoqun Peng,Zhufeng Chen,Xiaoyan Tang. Research Advances on Lectin Receptor-like Kinases in Plants [J]. Chinese Bulletin of Botany, 2020, 55(1): 96-105. |
| [8] | Qianqian Guo,Wenbin Zhou. Advances in the Mechanism Underlying Plant Response to Stress Combination [J]. Chinese Bulletin of Botany, 2019, 54(5): 662-673. |
| [9] | Jing Zhang,Suiwen Hou. Role of Post-translational Modification of Proteins in ABA Signaling Transduction [J]. Chinese Bulletin of Botany, 2019, 54(3): 300-315. |
| [10] | Hongyan Ren, Li Wang, Qingxiu Ma, Guang Wu. Progress in Biosynthetic Pathways of Brassinosteroids [J]. Chinese Bulletin of Botany, 2015, 50(6): 768-778. |
| [11] | Guizhi Zhang, Jishan Lin, Yanjie Li, Bingkai Hou. Research Advances in the Glycosylation of Plant Hormones [J]. Chinese Bulletin of Botany, 2014, 49(5): 515-523. |
| [12] | Panglian Xu;Mianwei Zeng;Lixia Huang;Chengwei Yang*. SUMOylation and Its Biological Function in Plants [J]. Chinese Bulletin of Botany, 2008, 25(05): 608-615. |
| [13] | Huixia Yang;Yiping Tong;Daowen Wang . Latest Advances in Understanding the Molecular Genetic Mechanism of Low Phosphate Responses in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2007, 24(06): 726-734. |
| [14] | Zhaoqing Chu;Li Li;Li Song;Hongwei Xue. Advances on Brassinosteroid Biosynthesis and Functions [J]. Chinese Bulletin of Botany, 2006, 23(5): 543-555. |
| [15] | Li Song;Li Li;Zhaoqin Chu;Hongwei Xue. Brassinosteroids Signal Transduction in Arabidopsis [J]. Chinese Bulletin of Botany, 2006, 23(5): 556-563. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||