

Chinese Bulletin of Botany ›› 2017, Vol. 52 ›› Issue (2): 188-201.DOI: 10.11983/CBB16054 cstr: 32102.14.CBB16054
Previous Articles Next Articles
Zheng Jun1, Qiao Ling1, Zhao Jiajia1, Qiao Linyi2, Zhang Shichang2, Chang Jianzhong2, Tang Caiguo3,*( ), Yang Sanwei1,*(
), Yang Sanwei1,*( )
)
Received:2016-03-18
Accepted:2016-08-04
Online:2017-03-01
Published:2017-04-05
Contact:
Tang Caiguo,Yang Sanwei
About author:# Co-first authors
Zheng Jun, Qiao Ling, Zhao Jiajia, Qiao Linyi, Zhang Shichang, Chang Jianzhong, Tang Caiguo, Yang Sanwei. Whole-genome Analysis of CCT Gene Family and Their Responses to Phytohormones in Aegilops tauschii[J]. Chinese Bulletin of Botany, 2017, 52(2): 188-201.
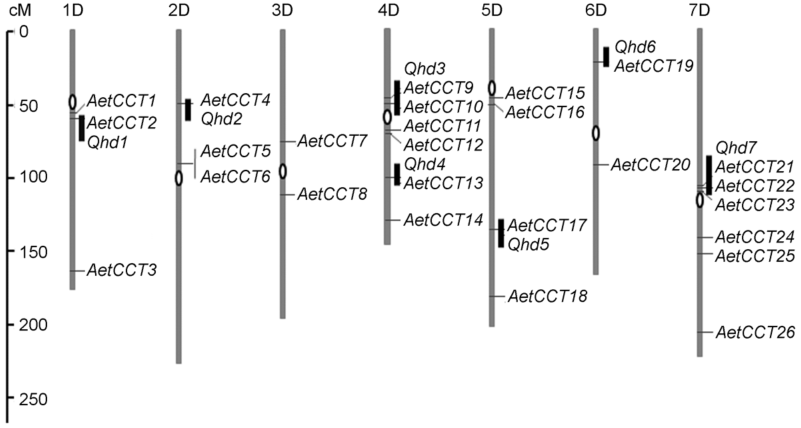
Figure 1 Chromosome mapping and QTL distribution of CCT genes in Aegilops tauschii genomeGenetic scale is indicated on the left side of chromosomes, the heading date QTLs are indicated on the right side of the chromosomes in black.
| Gene | Length (bp) | CDS | Scaffold | Location | Mapped seq. | Chromosome |
|---|---|---|---|---|---|---|
| AetCCT1 | 2211 | AEGTA18365 | Scaffold4528 | 49937-52147 | AT1D0317 | 1DL |
| AetCCT2 | 1836 | AEGTA21580 | Scaffold71509 | 42038-43873 | AT1D0523 | 1DL |
| AetCCT3 | 1333 | AEGTA27323 | Scaffold97182 | 45576-47039 | AT1D0969 | 1DL |
| AetCCT4 | 3096 | AEGTA10532 | Scaffold38896 | 10923-14018 | AT2D1124 | 2DS |
| AetCCT5 | 2241 | AEGTA13911 | Scaffold30755 | 94901-97141 | AT2D1581 | 2DS |
| AetCCT6 | 2157 | AEGTA00203 | Scaffold30755 | 129179-131557 | AT2D1581 | 2DS |
| AetCCT7 | 2226 | AEGTA31460 | Scaffold77907 | 6990-9215 | AT3D2607 | 3DS |
| AetCCT8 | 1134 | AEGTA06337 | Scaffold4745 | 59908-61041 | AT3D3130 | 3DL |
| AetCCT9 | 2692 | AEGTA28548 | Scaffold28581 | 76532-79223 | AT4D3632 | 4DS |
| AetCCT10 | 606 | AEGTA01709 | Scaffold50727 | 43106-43711 | AT4D3683 | 4DS |
| AetCCT11 | 3208 | AEGTA21446 | Scaffold108 | 140572-143968 | AT4D3728 | 4DL |
| AetCCT12 | 4521 | AEGTA31746 | Scaffold2864 | 19243-23723 | AT4D3886 | 4DL |
| AetCCT13 | 2234 | AEGTA18304 | Scaffold98624 | 23875-26108 | BE403305 | 4DL |
| AetCCT14 | 2878 | AEGTA13770 | Scaffold12030 | 253420-256287 | AT4D4194 | 4DL |
| AetCCT15 | 603 | AEGTA33063 | Scaffold24714 | 5731-6333 | AT5D4519 | 5DL |
| AetCCT16 | 1059 | AEGTA03508 | Scaffold185863 | 19051-20109 | AT5D4590 | 5DL |
| AetCCT17 | 2404 | AEGTA04421 | Scaffold53469 | 30253-32656 | AT5D5030 | 5DL |
| AetCCT18 | 738 | AEGTA32221 | Scaffold7166 | 12609-13346 | AT5D5201 | 5DL |
| AetCCT19 | 3597 | AEGTA05461 | Scaffold106936 | 10967-14734 | AT6D5284 | 6DS |
| AetCCT20 | 1188 | AEGTA21198 | Scaffold137524 | 2620-3965 | AT6D5798 | 6DL |
| AetCCT21 | 2031 | AEGTA15475 | Scaffold71269 | 16605-18635 | AT7D6397 | 7DS |
| AetCCT22 | 2781 | AEGTA31079 | Scaffold66553 | 48279-51059 | AT7D6416 | 7DS |
| AetCCT23 | 3075 | AEGTA08066 | Scaffold6305 | 52570-55644 | AT7D6716 | 7DS |
| AetCCT24 | 1521 | AEGTA13638 | Scaffold7100 | 34283-35803 | AT7D7002 | 7DL |
| AetCCT25 | 742 | AEGTA03492 | Scaffold116052 | 16157-16929 | AT7D7027 | 7DL |
| AetCCT26 | 2036 | AEGTA22574 | Scaffold16211 | 9228-11710 | AT7D7167 | 7DL |
Table 1 CCT gene family in Aegilops tauschii
| Gene | Length (bp) | CDS | Scaffold | Location | Mapped seq. | Chromosome |
|---|---|---|---|---|---|---|
| AetCCT1 | 2211 | AEGTA18365 | Scaffold4528 | 49937-52147 | AT1D0317 | 1DL |
| AetCCT2 | 1836 | AEGTA21580 | Scaffold71509 | 42038-43873 | AT1D0523 | 1DL |
| AetCCT3 | 1333 | AEGTA27323 | Scaffold97182 | 45576-47039 | AT1D0969 | 1DL |
| AetCCT4 | 3096 | AEGTA10532 | Scaffold38896 | 10923-14018 | AT2D1124 | 2DS |
| AetCCT5 | 2241 | AEGTA13911 | Scaffold30755 | 94901-97141 | AT2D1581 | 2DS |
| AetCCT6 | 2157 | AEGTA00203 | Scaffold30755 | 129179-131557 | AT2D1581 | 2DS |
| AetCCT7 | 2226 | AEGTA31460 | Scaffold77907 | 6990-9215 | AT3D2607 | 3DS |
| AetCCT8 | 1134 | AEGTA06337 | Scaffold4745 | 59908-61041 | AT3D3130 | 3DL |
| AetCCT9 | 2692 | AEGTA28548 | Scaffold28581 | 76532-79223 | AT4D3632 | 4DS |
| AetCCT10 | 606 | AEGTA01709 | Scaffold50727 | 43106-43711 | AT4D3683 | 4DS |
| AetCCT11 | 3208 | AEGTA21446 | Scaffold108 | 140572-143968 | AT4D3728 | 4DL |
| AetCCT12 | 4521 | AEGTA31746 | Scaffold2864 | 19243-23723 | AT4D3886 | 4DL |
| AetCCT13 | 2234 | AEGTA18304 | Scaffold98624 | 23875-26108 | BE403305 | 4DL |
| AetCCT14 | 2878 | AEGTA13770 | Scaffold12030 | 253420-256287 | AT4D4194 | 4DL |
| AetCCT15 | 603 | AEGTA33063 | Scaffold24714 | 5731-6333 | AT5D4519 | 5DL |
| AetCCT16 | 1059 | AEGTA03508 | Scaffold185863 | 19051-20109 | AT5D4590 | 5DL |
| AetCCT17 | 2404 | AEGTA04421 | Scaffold53469 | 30253-32656 | AT5D5030 | 5DL |
| AetCCT18 | 738 | AEGTA32221 | Scaffold7166 | 12609-13346 | AT5D5201 | 5DL |
| AetCCT19 | 3597 | AEGTA05461 | Scaffold106936 | 10967-14734 | AT6D5284 | 6DS |
| AetCCT20 | 1188 | AEGTA21198 | Scaffold137524 | 2620-3965 | AT6D5798 | 6DL |
| AetCCT21 | 2031 | AEGTA15475 | Scaffold71269 | 16605-18635 | AT7D6397 | 7DS |
| AetCCT22 | 2781 | AEGTA31079 | Scaffold66553 | 48279-51059 | AT7D6416 | 7DS |
| AetCCT23 | 3075 | AEGTA08066 | Scaffold6305 | 52570-55644 | AT7D6716 | 7DS |
| AetCCT24 | 1521 | AEGTA13638 | Scaffold7100 | 34283-35803 | AT7D7002 | 7DL |
| AetCCT25 | 742 | AEGTA03492 | Scaffold116052 | 16157-16929 | AT7D7027 | 7DL |
| AetCCT26 | 2036 | AEGTA22574 | Scaffold16211 | 9228-11710 | AT7D7167 | 7DL |
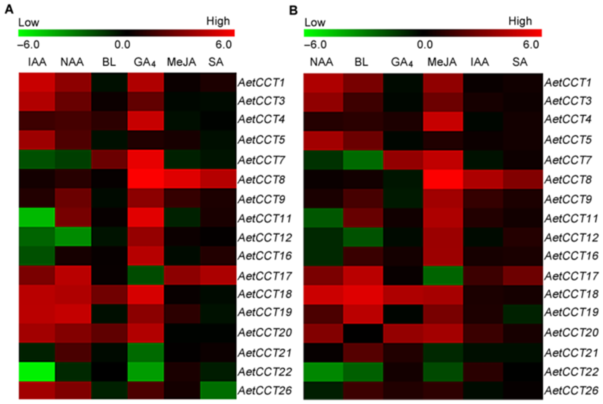
Figure 8 Hierarchical clustering of CCT domain genes under different phytohormones treatments in Aegilops tauschii(A) Phytohormones treatment for 24 h; (B) Phytohormones treatment for 72 h
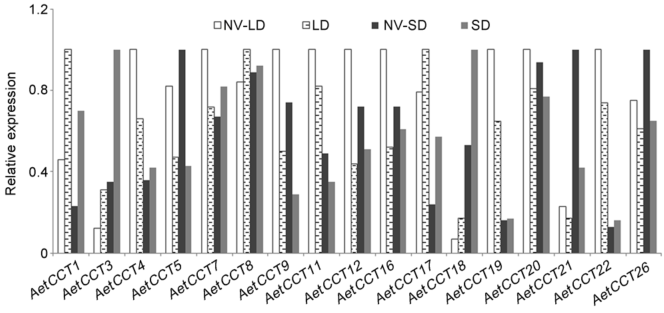
Figure 9 Expression of CCT domain genes under different light treatments in Aegilops tauschiiNV-LD: Long-day condition without vernalization (16 h light/8 h dark); NV-SD: Short-day condition without vernalization (8 h light/16 h dark); LD: Long-day condition after vernalization (16 h light/8 h dark); SD: Short-day condition after vernalization (8 h light/16 h dark).
| [1] | 陈华夏, 申国境, 王磊, 邢永忠 (2010). 4个物种CCT结构域基因家族的序列进化分析. 华中农业大学学报 29, 669-676. |
| [2] | Ando E, Ohnishi M, Wang Y, Matsushita T, Watanabe A, Hayashi Y, Fujii M, Ma JF, Inoue S, Kinoshita T (2013). TWIN SISTER OF FT,GIGANTEA, and CONSTANS ha- ve a positive but indirect effect on blue light-induced stom- atal opening in Arabidopsis. Plant Physiol 162, 1529-1538. |
| [3] | Cho LH, Choi H, An G (2010). OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB.Plant J 63, 8-30. |
| [4] | Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W (2007). Control of flowering time in temperate cereals: genes, domestication and sustainable productivity.J Exp Bot 58, 1231-1244. |
| [5] | Cockram J, Thiel T, Steuernagel B, Stein N, Taudien S, Bailey PC, Sullivan DM (2012). Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae.PLoS One 7, e45307. |
| [6] | Dennis ES, Peacock WJ (2009). Vernalization in cereals.J Biol 8, 57. |
| [7] | Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controlsFT-like gene expression indepen- dently of Hd1. Genes Dev 18, 926-936. |
| [8] | Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma- Bognár L, Nagy F, Millar AJ, Amasino RM (2002). TheELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana.Nature 419, 74-77. |
| [9] | Gao H, Zheng XM, Fei GL, Chen J, Jin MN, Ren YL, Wu WX, Zhou KN, Sheng PK, Zhou F, Jiang L, Wang J, Zhang X, Guo XP, Wang JL, Cheng ZJ, Wu CY, Wang HY, Wan JM (2013). Ehd4 encodes a novel andOryza- genus-specific regulator of photoperiodic flowering in rice. PLoS Genet 9, e1003281. |
| [10] | Grasser KD (2005). Emerging role for transcript elongation in plant development.Trends Plant Sci 10, 484-490. |
| [11] | Harmon F, Imaizumi T, Gray WM (2008). CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock.Plant J 55, 568-579. |
| [12] | Hicks KA, Albertson TM, Wagner DR (2001). EARLY FLO- WERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis.Plant Cell 13, 1281-1292. |
| [13] | Hsu CY, Adams JP, No K, Liang H, Meilan R, Pechanova O, Barakat A, Carlson JE, Page GP, Yuceer C (2012). Overexpression ofCONSTANS homologs CO1 and CO2 fails to alter normal reproductive onset and fall bud set in woody perennial poplar. PLoS One 7, e45448. |
| [14] | Kim J, Kim Y, Yeom M, Kim JH, Nam HG (2008). FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20, 307-319. |
| [15] | Kim SK, Park HY, Jang YH, Lee JH, Kim JK (2013). The sequence variation responsible for the functional difference between the CONSTANS protein, and the CONSTANS-like COL1 and COL2 proteins, resides mostly in the region encoded by their first exons. Plant Sci 199-200, 71-78. |
| [16] | Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J, Rademacher EH, Moller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D (2011). A cellular expression map of the ArabidopsisAUXIN RESPONSE FACTOR gene family. Plant J 68, 597-606. |
| [17] | Lolas IB, Himanen K, Gronlund JT, Lynggaard C, Houben A, Melzer M, Van Lijsebettens M, Grasser KD (2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2.Plant J 61, 686-697. |
| [18] | Panda S, Poirier GG, Kay SA (2002). Tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator.Dev Cell 3, 51-61. |
| [19] | Putterill J, Robson F, Lee K, Simon R, Coupland G (1995). TheCONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847-857. |
| [20] | Riboni M, Galbiati M, Tonelli C, Conti L (2013). GIGANTEA enables drought escape response via abscisic acid- dependent activation of the florigens andSUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol 162, 1706-1719. |
| [21] | Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, Matsubara K, Hori K, Strader LC, Chen GL, Bartel B (2010). Ethylene directs auxin to control root cell expansion.Plant J 64, 874-884. |
| [22] | Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000). Cloning of the Arabidopsis clock geneTOC1, an autoregulatory response regulator homolog. Science 289, 768-771. |
| [23] | Trevaskis B, Hemming MN, Dennis ES, Peacock WJ (2007). The molecular basis of vernalization-induced flo- wering in cereals.Trends Plant Sci 12, 352-357. |
| [24] | Vanneste S, Friml J (2009). Auxin: a trigger for change in plant development.Cell 136, 1005-1016. |
| [25] | Wu F, Price B, Haider W, Seufferheld G, Nelson R, Hanzawa Y (2014). Functional and evolutionary characterization of theCONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS One 9, e85754. |
| [26] | Wu WX, Zheng XM, Lu GW, Zhong ZZ, Gao H, Chen LP, Wu CY, Wang HJ, Wang Q, Zhou KN, Wang JL, Wu FQ, Zhang X, Guo XP, Cheng ZJ, Lei CL, Lin QB, Jiang L, Wang HY, Ge S, Wan JM (2013). Association of functional nucleotide polymorphisms atDTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA 110, 2775-2780. |
| [27] | Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, Gao C, Chong K (2014). O-GlcNAc- mediated interaction between ?VER2 and ?TaGRP2 elicits ?TaVRN1 mRNA accumulation during vernalization in win- ter wheat. Nat Commun 5, 4572-4578. |
| [28] | Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, Zhang QF (2008). Natural variation inGhd7 is an important regulator of hea- ding date and yield potential in rice. Nat Genet 40, 761-767. |
| [29] | Yan L, Fu DL, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006). The wheat and barley vernalization geneVRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103, 19581-19586. |
| [30] | Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakri- shna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004). The wheatVRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640-1644. |
| [31] | Yang Q, Li Z, Li WQ, Ku LX, Wang C, Ye JR, Li K, Yang N, Li YP, Zhong T, Li JS, Chen YH, Yan JB, Yang XH, Xu ML (2013). CACTA-like transposable element inZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci USA 110, 16969-16974. |
| [32] | Yano M, Inoue H, Tanisaka T (2012). Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor geneGhd7 under both short- and long-day conditions. Plant Cell Physiol 53, 717-728. |
| [33] | Zhang L, Li QP, Dong HJ, He Q, Liang LW, Tan C, Han ZM, Yao W, Li GW, Zhao H, Xie WB, Xin YZ (2015). Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci Rep 5, 7663. |
| [34] | Zheng J, Liu H, Wang YQ, Wang LF, Chang XP, Jing RL, Hao CY, Zhang XY (2014). TaTEF-7A, a transcript elongation factor gene, influences yield-related traits in bread wheat (Triticum aestivum L). J Exp Bot 65, 5351-5365. |
| [1] | Keyi Wu, Wenda Ruan, Difeng Zhou, Qingchen Chen, Chengyun Zhang, Xinyuan Pan, Shang Yu, Yang Liu, Rongbo Xiao. Syllable clustering analysis-based passive acoustic monitoring technology and its application in bird monitoring [J]. Biodiv Sci, 2023, 31(1): 22370-. |
| [2] | Shaoshuai Yu, Caili Lin, Shengjie Wang, Wenxin Zhang, Guozhong Tian. Structures of the tuf gene and its upstream part genes and characteristic analysis of conserved regions and activity from related gene promoters of a phytoplasma [J]. Biodiv Sci, 2018, 26(7): 738-748. |
| [3] | Shaoshuai Yu, Qicong Xu, Caili Lin, Shengjie Wang, Guozhong Tian. Genetic diversity of phytoplasmas: research status and prospects [J]. Biodiv Sci, 2016, 24(2): 205-215. |
| [4] | Li-Long WANG, Liang WANG, Li-Fang ZHANG, Yu-Yang LIU, Shi-Jian XU. Structure and dynamic characteristics of Gymnocarpos przewalskii in different habitats [J]. Chin J Plant Ecol, 2015, 39(10): 980-989. |
| [5] | Xi Zhang, Xiaogai Hou, Dalong Guo, Chengwei Song, Yabin Duan. iPBS-PCR Used for Cloning and Analysis of Long Terminal Repeat Transposons in Tree Peony (Paeonia) [J]. Chinese Bulletin of Botany, 2014, 49(3): 322-330. |
| [6] | Gen Zhang, Yilong Xi, Yinghao Xue, Xin Hu, Xianling Xiang, Xinli Wen. Effects of coal ash pollution on the genetic diversity of Brachionus calyciflorus based on rDNA ITS sequences [J]. Biodiv Sci, 2010, 18(3): 241-250. |
| [7] | Ge Yao;Shulian Xie. Progress in Molecular Systematics of Batrachospermales [J]. Chinese Bulletin of Botany, 2007, 24(02): 141-146. |
| [8] | LI Chun-Xiang YANG Qun. Direct Sequencing of PCR Products or Sequencing by Cloning PCR Products—Method of Determining Internal Transcribed Spacer Sequences of Nuclear Ribosomal DNA inAthrotaxis [J]. Chinese Bulletin of Botany, 2002, 19(06): 698-704. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||