

Chinese Bulletin of Botany ›› 2024, Vol. 59 ›› Issue (5): 774-782.DOI: 10.11983/CBB24107 cstr: 32102.14.CBB24107
• INVITED PROTOCOL • Previous Articles Next Articles
Chunjiao Xia, Yunguang Li, Shu Xia, Wei Pang, Chunli Chen*( )
)
Received:2024-07-18
Accepted:2024-08-20
Online:2024-09-10
Published:2024-08-22
Contact:
Chunli Chen
Chunjiao Xia, Yunguang Li, Shu Xia, Wei Pang, Chunli Chen. Flow Cytometric Analysis and Sorting in Plant Genomics[J]. Chinese Bulletin of Botany, 2024, 59(5): 774-782.
| Names | Components | Applications |
|---|---|---|
| Galbraith’s (Galbraith et al., | 45 mmol∙L-1 MgCl2, 30 mmol∙L-1 sodium citrate, 20 mmol∙L-1 MOPS, 0.1% (v/v) TritonX-100, pH7.0 | Arabidopsis thaliana, Glycine max |
| LB01 (Dpooležel et al., | 15 mmol∙L-1 Tris, 2 mmol∙L-1 Na2EDTA, 0.5 mmol∙L-1 spermine, 80 mmol∙L-1 KCl, 20 mmol∙L-1 NaCl, 0.1% (v/v) TritonX-100, 15 mmol∙L-1 β-mercaptoethanol, pH7.0-8.0 | Oryza sativa, Chrysanthemum indicum, Solanum lycopersicum |
| Otto’s (Otto, | OTTO I: 100 mmol∙L-1 citric acid, 0.5% (v/v) Tween-20, pH2.0- 3.0; OTTO II: 400 mmol∙L-1 Na2HPO4·12H2O, pH8.0-9.0 | Ranunculus japonicus, O. sativa |
| Tris-MgCl2 (Pfosser et al., | 200 mmol∙L-1 Tris, 4 mmol∙L-1 MgCl2·6H2O, 0.5% (v/v) TritonX- 100, pH7.5 | Centaurea cyanus, Celtis au- stralis |
| GPB (Loureiro et al., | 0.5 mmol∙L-1 spermine, 30 mmol∙L-1 sodium citrate, 20 mmol∙L-1 MOPS, 80 mmol∙L-1 KCl, 20 mmol∙L-1 NaCl, 0.5% (v/v) TritonX- 100, pH7.0 | O. sativa, Actinidia chinensis |
| WPB (Loureiro et al., | 0.2 mol∙L-1 Tris-HCl, 4 mmol∙L-1 MgCl2∙6H2O, 2 mmol∙L-1 EDTA Na2·2H2O, 86 mmol∙L-1 NaCl, 10 mmol∙L-1 sodium pyrosulfite, 1% PVP-10, 1% (v/v) TritonX-100, pH7.5 | Vitis vinifera, Quercus robur |
| PVPK12-mGB2 (Zhang and Feng, | 30 mmol∙L-1 sodium citrate, 45 mmol∙L-1 MgCl2, 20 mmol∙L-1 MO- PS, 20 mmol∙L-1 NaCl, 20 mmol∙L-1 EDTA Na2·2H2O, 0.1% (v/v) TritonX-100, 0.5% (v/v) Tween-20, 10 μL·mL-1 β-mercaptoethanol, 1%-2% PVPK12, pH7.0 | Silicone fast-drying plant ma- terials |
Table 1 Several representative plant cell nucleus dissociation buffers
| Names | Components | Applications |
|---|---|---|
| Galbraith’s (Galbraith et al., | 45 mmol∙L-1 MgCl2, 30 mmol∙L-1 sodium citrate, 20 mmol∙L-1 MOPS, 0.1% (v/v) TritonX-100, pH7.0 | Arabidopsis thaliana, Glycine max |
| LB01 (Dpooležel et al., | 15 mmol∙L-1 Tris, 2 mmol∙L-1 Na2EDTA, 0.5 mmol∙L-1 spermine, 80 mmol∙L-1 KCl, 20 mmol∙L-1 NaCl, 0.1% (v/v) TritonX-100, 15 mmol∙L-1 β-mercaptoethanol, pH7.0-8.0 | Oryza sativa, Chrysanthemum indicum, Solanum lycopersicum |
| Otto’s (Otto, | OTTO I: 100 mmol∙L-1 citric acid, 0.5% (v/v) Tween-20, pH2.0- 3.0; OTTO II: 400 mmol∙L-1 Na2HPO4·12H2O, pH8.0-9.0 | Ranunculus japonicus, O. sativa |
| Tris-MgCl2 (Pfosser et al., | 200 mmol∙L-1 Tris, 4 mmol∙L-1 MgCl2·6H2O, 0.5% (v/v) TritonX- 100, pH7.5 | Centaurea cyanus, Celtis au- stralis |
| GPB (Loureiro et al., | 0.5 mmol∙L-1 spermine, 30 mmol∙L-1 sodium citrate, 20 mmol∙L-1 MOPS, 80 mmol∙L-1 KCl, 20 mmol∙L-1 NaCl, 0.5% (v/v) TritonX- 100, pH7.0 | O. sativa, Actinidia chinensis |
| WPB (Loureiro et al., | 0.2 mol∙L-1 Tris-HCl, 4 mmol∙L-1 MgCl2∙6H2O, 2 mmol∙L-1 EDTA Na2·2H2O, 86 mmol∙L-1 NaCl, 10 mmol∙L-1 sodium pyrosulfite, 1% PVP-10, 1% (v/v) TritonX-100, pH7.5 | Vitis vinifera, Quercus robur |
| PVPK12-mGB2 (Zhang and Feng, | 30 mmol∙L-1 sodium citrate, 45 mmol∙L-1 MgCl2, 20 mmol∙L-1 MO- PS, 20 mmol∙L-1 NaCl, 20 mmol∙L-1 EDTA Na2·2H2O, 0.1% (v/v) TritonX-100, 0.5% (v/v) Tween-20, 10 μL·mL-1 β-mercaptoethanol, 1%-2% PVPK12, pH7.0 | Silicone fast-drying plant ma- terials |
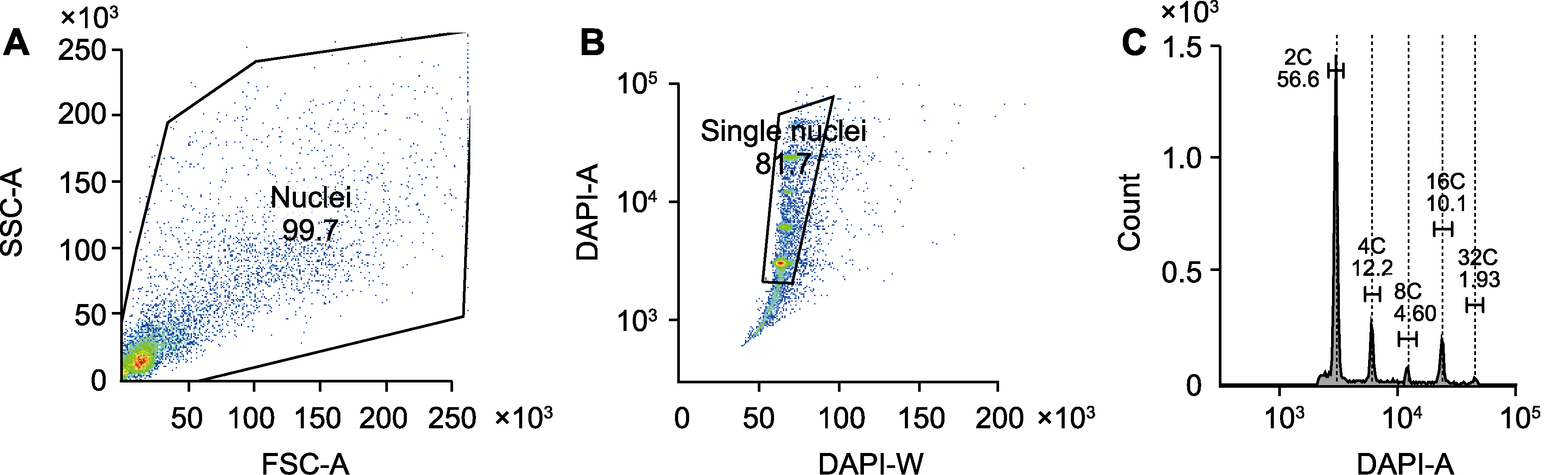
Figure 2 Analysis of different DNA ploidies in nuclei of soybean nodule cells (A) Total nucleus gate was set by FSC-A and SSC-A dual parameters; (B) Single nuclei gate was set by DAPI-W and DAPI-A; (C) Nuclei gate for different DNA ploidies was set by DAPI-A histogram. SSC and FSC are the same as shown in Figure 1. DAPI: 4,6-diamidino-2-phenylindole
| [1] | Bressan D, Battistoni G, Hannon GJ (2023). The dawn of spatial omics. Science 381, eabq4964. |
| [2] | Büscher M (2019). Flow cytometry instrumentation—an overview. Curr Protoc Cytom doi: 10.1002/cpcy.52. |
| [3] | Cápal P, Said M, Molnár I, Doležel J (2023). Flow cytometric analysis and sorting of plant chromosomes. In: Heitkam T, Garcia S, eds. Plant Cytogenetics and Cytogenomics. New York: Humana. pp. 177-200. |
| [4] | Decaestecker W, Bollier N, Buono RA, Nowack MK, Jacobs TB (2022). Protoplast preparation and fluorescence- activated cell sorting for the evaluation of targeted mutagenesis in plant cells. In: Wang K, Zhang F, eds. Protoplast Technology. New York: Humana. pp. 205-221. |
| [5] | Doležel J, Číhalíková J, Lucretti S (1992). A high-yield procedure for isolation of metaphase chromosomes from root tips of Vicia faba L. Planta 188, 93-98. |
| [6] | Doležel J, Göhde W (1995). Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry 19, 103-106. |
| [7] | Doležel J, Greilhuber J, Suda J (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2, 2233-2244. |
| [8] | Doležel J, Lucretti S, Molnár I, Cápal P, Giorgi D (2021). Chromosome analysis and sorting. Cytometry A 99, 328- 342. |
| [9] | Dpooležel J, Binarová P, Lcretti S (1989). Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant 31, 113-120. |
| [10] | Fan W, Xia CJ, Wang SX, Liu J, Deng LJ, Sun SY, Wang XL (2022). Rhizobial infection of 4C cells triggers their endoreduplication during symbiotic nodule development in soybean. New Phytol 234, 1018-1030. |
| [11] | Galbraith DW, Afonso CL, Harkins KR (1984). Flow sorting and culture of protoplasts: conditions for high-frequency recovery, growth and morphogenesis from sorted protoplasts of suspension cultures of nicotiana. Plant Cell Rep 3, 151-155. |
| [12] | Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049-1051. |
| [13] | Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O'Shaughnessy AL, Lambert GM, Araúzo- Bravo MJ, Lee J, Fishman M, Robbins GE, Lin XY, Venepally P, Badger JH, Galbraith DW, Gage FH, Lasken RS (2013). RNA-sequencing from single nuclei. Proc Natl Acad Sci USA 110, 19802-19807. |
| [14] | Guedes JG, Guimaraes AL, Carqueijeiro I, Gardner R, Bispo C, Sottomayor M (2022). Isolation of specialized plant cells by fluorescence-activated cell sorting. In: Fett-Neto AG,ed. Plant Secondary Metabolism Engineering. New York: Humana. pp. 193-200. |
| [15] | Guillotin B, Rahni R, Passalacqua M, Mohammed MA, Xu XS, Raju SK, Ramírez CO, Jackson D, Groen SC, Gillis J, Birnbaum KD (2023). A pan-grass transcriptome reveals patterns of cellular divergence in crops. Nature 617, 785-791. |
| [16] | Hang HY, Liu CC, Ren DD (2019). Development, application and prospection of flow cytometry. China Biotechnol 39(9), 68-83. (in Chinese) |
| 杭海英, 刘春春, 任丹丹 (2019). 流式细胞术的发展、应用及前景. 中国生物工程杂志 39(9), 68-83. | |
| [17] | Kamentsky LA, Melamed MR, Derman H (1965). Spectrophotometer: new instrument for ultrarapid cell analysis. Science 150, 630-631. |
| [18] | Kawakatsu T, Stuart T, Valdes M, Breakfield N, Schmitz RJ, Nery JR, Urich MA, Han XW, Lister R, Benfey PN, Ecker JR (2016). Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat Plants 2, 16058. |
| [19] | Li F, Hu Y, Wang F, Zhang Z, Liu XL, Bai SL, He YK (2010). Flow cytometry sorting of early developmental non- hair cells in roots of Arabidopsis thaliana. Chin Bull Bot 45, 460-465. (in Chinese) |
| 李斐, 胡勇, 王帆, 张珍, 刘祥林, 白素兰, 何奕昆 (2010). 利用流式细胞仪分选拟南芥根尖发育早期非根毛细胞. 植物学报 45, 460-465. | |
| [20] | Lin CS, Hsu CT, Yang LH, Lee LY, Fu JY, Cheng QW, Wu FH, Hsiao HCW, Zhang YS, Zhang R, Chang WJ, Yu CT, Wang W, Liao LJ, Gelvin SB, Shih MC (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 16, 1295-1310. |
| [21] | Loureiro J, Rodriguez E, Doležel J, Santos C (2007). Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot 100, 875-888. |
| [22] | Marx V (2016). Plants: a tool box of cell-based assays. Nat Methods 13, 551-554. |
| [23] | Mayer KFX, Martis M, Hedley PE, Šimková H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubaláková M, Suchánková P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Doležel J, Waugh R, Stein N (2011). Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23, 1249-1263. |
| [24] | Nitta N, Sugimura T, Isozaki A, Mikami H, Hiraki K, Sakuma S, Iino T, Arai F, Endo T, Fujiwaki Y, Fukuzawa H, Hase M, Hayakawa T, Hiramatsu K, Hoshino Y, Inaba M, Ito T, Karakawa H, Kasai Y, Koizumi K, Lee S, Lei C, Li M, Maeno T, Matsusaka S, Murakami D, Nakagawa A, Oguchi Y, Oikawa M, Ota T, Shiba K, Shintaku H, Shirasaki Y, Suga K, Suzuki Y, Suzuki N, Tanaka Y, Tezuka H, Toyokawa C, Yalikun Y, Yamada M, Yamagishi M, Yamano T, Yasumoto A, Yatomi Y, Yazawa M, Di Carlo D, Hosokawa Y, Uemura S, Ozeki Y, Goda K (2018). Intelligent image-activated cell sorting. Cell 175, 266-276. |
| [25] | Ortiz-Ramírez C, Arevalo ED, Xu XS, Jackson DP, Birnbaum KD (2018). An efficient cell sorting protocol for maize protoplasts. Curr Protoc Plant Biol 3, e20072. |
| [26] | Otto FJ (1992). Preparation and staining of cells for high- resolution DNA analysis. In: Radbruch A, ed. Flow Cytometry and Cell Sorting. Berlin: Springer. pp. 65-68. |
| [27] | Petrovská B, Jeřábková H, Chamrád I, Vrána J, Lenobel R, Uřinovská J, Šebela M, Doležel J (2014). Proteomic analysis of barley cell nuclei purified by flow sorting. Cytogenet Genome Res 143, 78-86. |
| [28] | Pfosser M, Heberle-Bors E, Amon A, Lelley T (1995). Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat- rye addition lines. Cytometry 21, 387-393. |
| [29] | Reichard A, Asosingh K (2019). Best practices for preparing a single cell suspension from solid tissues for flow cytometry. Cytometry A 95, 219-226. |
| [30] | Šafář J, Bartoš J, Janda J, Bellec A, Kubaláková M, Valárik M, Pateyron S, Weiserová J, Tušková R, Číhalíková J, Vrána J, Šimková H, Faivre-Rampant P, Sourdille P, Caboche M, Bernard M, Doležel J, Chalhoub B (2004). Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J 39, 960-968. |
| [31] | Said M, Kubaláková M, Karafiátová M, Molnár I, Doležel J, Vrána J (2019). Dissecting the complex genome of crested wheatgrass by chromosome flow sorting. Plant Genome 12, 180096. |
| [32] | Schraivogel D, Kuhn TM, Rauscher B, Rodríguez-Martínez M, Paulsen M, Owsley K, Middlebrook A, Tischer C, Ramasz B, Ordoñez-Rueda D, Dees M, Cuylen- Haering S, Diebold E, Steinmetz LM (2022). High-speed fluorescence image-enabled cell sorting. Science 375, 315- 320. |
| [33] | Shapiro HM (2003). Practical Flow Cytometry, 4th edn. New York: Wiley-Liss. pp. 73-100. |
| [34] | Sliwinska E, Loureiro J, Leitch IJ, Šmarda P, Bainard J, Bureš P, Chumová Z, Horová L, Koutecký P, Lučanová M, Trávníček P, Galbraith DW (2022). Application-based guidelines for best practices in plant flow cytometry. Cytometry A 101, 749-781. |
| [35] | Smith JP, Sheffield NC (2020). Analytical approaches for ATAC-seq data analysis. Curr Protoc Hum Genet 106, e101. |
| [36] | Sun GL, Xia MZ, Li JP, Ma W, Li QZ, Xie JJ, Bai SL, Fang SS, Sun T, Feng XL, Guo GH, Niu YL, Hou JY, Ye WL, Ma JC, Guo SY, Wang HL, Long Y, Zhang XB, Zhang JL, Zhou H, Li BZ, Liu J, Zou CS, Wang H, Huang JL, Galbraith DW, Song CP (2022). The maize single-nucleus transcriptome comprehensively describes signaling networks governing movement and development of grass stomata. Plant Cell 34, 1890-1911. |
| [37] | Taher TEI (2017). Monitoring promoter activity by flow cytometry. In: Gould D, ed. Mammalian Synthetic Promoters. New York: Humana. pp. 65-73. |
| [38] | The International Wheat Genome Sequencing Consortium IWGSC (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. |
| [39] | Xu XS, Crow M, Rice BR, Li F, Harris B, Liu L, Demesa-Arevalo E, Lu ZF, Wang LY, Fox N, Wang XF, Drenkow J, Luo AD, Char SN, Yang B, Sylvester AW, Gingeras TR, Schmitz RJ, Ware D, Lipka AE, Gillis J, Jackson D (2021). Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev Cell 56, 557-568. |
| [40] | Zhang JD, Feng M (2023). A plant sample optimal pretreatment for flow cytometric analysis. Chin Bull Bot 58, 285-297. (in Chinese) |
| 张晋丹, 冯旻 (2023). 一种提升流式细胞术分析效果的前处理方法. 植物学报 58, 285-297. | |
| [41] | Zhu T, Liao KY, Zhou RF, Xia CJ, Xie WB (2020). ATAC- seq with unique molecular identifiers improves quantification and footprinting. Commun Biol 3, 675. |
| [42] | Zhu T, Xia CJ, Yu RR, Zhou XK, Xu XB, Wang L, Zong ZX, Yang JJ, Liu YM, Ming LC, You YX, Chen DJ, Xie WB (2024). Comprehensive mapping and modelling of the rice regulome landscape unveils the regulatory architecture underlying complex traits. Nat Commun 15, 6562. |
| [1] | Lu Zi-Jia, Wang Tian-Rui, Zheng Si-Si, Cao Jian-Guo, Kozlowski Gregor, Song Yi-Gang. Environmental adaptive genetic variation and genetic vulnerability of relict plant Pterocarya hupehensis [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [2] | ZHANG Zi-Rui, Zhou Jing, HU Yan-Ping, Liang Shuang, MA Yong-Peng, CHEN Wei-Le. Root-associated Fungal Communities of the Critically Endangered Plant Pinus Squamata [J]. , 2025, 49(濒危植物的保护与恢复): 0-. |
| [3] | Gan Xie, Jing Xuan, Qidi Fu, Ze Wei, Kai Xue, Hairui Luo, Jixi Gao, Min Li. Establishing an intelligent identification model for unmanned aerial vehicle surveys of grassland plant diversity [J]. Biodiv Sci, 2025, 33(4): 24236-. |
| [4] | Tao Xie, Yifan Zhang, Yunhui Liu, Huiyu You, Jibenben Xia, Rong Ma, Chunni Zhang, Xuejun Hua. Research progress of iron-sulfur cluster synthesis system and regulation in plant mitochondria [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [5] | JIANG Xiao-Yu, YU Xin-Miao, LIAO Qin, ZHANG Jin-Wei, WU Xue-Feng, WANG Xu, PAN Jun-Tong, WANG Jun-Feng, MU Chun-Sheng, SHI Yu-Jie. Studies on the emission of nitrous oxide from terrestrial plants [J]. Chin J Plant Ecol, 2025, 49(4): 513-525. |
| [6] | Xiong Lianglin, Liang Guolu, Guo Qigao, Jing Danlong. Advances in the Regulation of Alternative Splicing of Genes in Plants in Response to Abiotic Stress [J]. Chinese Bulletin of Botany, 2025, 60(3): 435-448. |
| [7] | Zhang Chan, Zhao Suya, Zhang Xinran, Wang Yifan, Wang Linlin. Impacts of alien pollinators on native plant‒pollinator interactions [J]. Biodiv Sci, 2025, 33(2): 24443-. |
| [8] | Yaping Wang, Wenquan Bao, Yu’e Bai. Advances in the Application of Single-cell Transcriptomics in Plant Growth, Development and Stress Response [J]. Chinese Bulletin of Botany, 2025, 60(1): 101-113. |
| [9] | Ruoyue Li, Xiaochao Yang, Zhanqing Hao, Shihong Jia. The intensity of heat waves and insect herbivory on campus plants and their relationship with leaf functional traits [J]. Biodiv Sci, 2025, 33(1): 24283-. |
| [10] | Jing Xuan, Qidi Fu, Gan Xie, Kai Xue, Hairui Luo, Ze Wei, Mingyue Zhao, Liang Zhi, Huawei Wan, Jixi Gao, Min Li. An Artificial Intelligence Model for Identifying Grassland Plants in Northern China [J]. Chinese Bulletin of Botany, 2025, 60(1): 74-80. |
| [11] | Yan Deng, Limin Lu, Qiang Zhang, Zhiduan Chen, Haihua Hu. A Comprehensive Evaluation of the Plastid DNA Data Gaps of Vascular Plants in Species and Geographic Area [J]. Chinese Bulletin of Botany, 2025, 60(1): 1-16. |
| [12] | Xiangtan Yao, Xinyi Zhang, Yang Chen, Ye Yuan, Wangda Cheng, Tianrui Wang, Yingxiong Qiu. Genomic resequencing reveals the genetic diversity of the cultivated water caltrop, and the origin and domestication of ‘Nanhuling’ [J]. Biodiv Sci, 2024, 32(9): 24212-. |
| [13] | WANG Yi-Tong, Yeerjiang BAIKETUERHAN, LIAO Dan, WANG Juan. Correlation between elemental biometric characteristics and sexual dimorphism in leaves of dioecious Acer barbinerve at different growth stages [J]. Chin J Plant Ecol, 2024, 48(6): 760-769. |
| [14] | QU Ze-Kun, ZHU Li-Qin, JIANG Qi, WANG Xiao-Hong, YAO Xiao-Dong, CAI Shi-Feng, LUO Su-Zhen, sCHEN Guang-Shui. Nutrient foraging strategies of arbuscular mycorrhizal tree species in a subtropical evergreen broadleaf forest and their relationship with fine root morphology [J]. Chin J Plant Ecol, 2024, 48(4): 416-427. |
| [15] | Lansha Luo, Wenpei Song, Qingzhu Hua, Dawei Li, Hong Liang, Xianzhi Zhang. Research Progress on Plant Sex-determination Genes and Their Epigenetic Regulation [J]. Chinese Bulletin of Botany, 2024, 59(2): 278-290. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||