

Chinese Bulletin of Botany ›› 2023, Vol. 58 ›› Issue (4): 602-611.DOI: 10.11983/CBB22091 cstr: 32102.14.CBB22091
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Shengyu Liu, Xiaobin Liu, Jiafu Zhu, Jing Su, Zhicheng Dong, Min Liu( )
)
Received:2022-04-29
Accepted:2022-09-07
Online:2023-07-01
Published:2022-09-30
Contact:
*E-mail: minl@gzhu.edu.cn
Shengyu Liu, Xiaobin Liu, Jiafu Zhu, Jing Su, Zhicheng Dong, Min Liu. Optimization and Evaluation of Tn5 Transposase Fusion Protein in CUT&Tag[J]. Chinese Bulletin of Botany, 2023, 58(4): 602-611.
| Buffer | Component | Concentration | Buffer | Component | Concentration |
|---|---|---|---|---|---|
| HEGX buffer | NaCl | 0.8 mol·L-1 | HBC buffer | Tris-HCl, pH7.5 | 20 mmol·L-1 |
| Glycerol | 10% | Sucrose | 352 mmol·L-1 | ||
| TritonX-100 | 0.2% | MgCl2 | 8 mmol·L-1 | ||
| HEPES-KOH, pH7.2 | 20 mmol·L-1 | TritonX-100 | 0.08% | ||
| EDTA | 1 mmol·L-1 | β-mercaptoethanol | 8 mmol·L-1 | ||
| Storage buffer | NaCl | 0.8 mol·L-1 | Glycerol | 20% | |
| Glycerol | 10% | 10× binding buffer | HEPES-KOH, pH8.0 | 20 mmol·L-1 | |
| Tris-HCl, pH7.5 | 50 mmol·L-1 | KCl | 100 mmol·L-1 | ||
| EDTA | 0.2 mmol·L-1 | CaCl2 | 10 mmol·L-1 | ||
| DTT | 2 mmol·L-1 | MnCl2 | 10 mmol·L-1 | ||
| Annealing buffer | Tris-HCl, pH7.0 | 5 mmol·L-1 | Spermidine | 5 mmol·L-1 | |
| EDTA | 0.5 mmol·L-1 | Blocking buffer | HEPES-KOH, pH7.5 | 20 mmol·L-1 | |
| NaCl | 50 mmol·L-1 | NaCl | 150 mmol·L-1 | ||
| 5× Tagmentation buffer | TAPS-NaOH, pH8.5 | 50 mmol·L-1 | Spermidine | 0.5 mmol·L-1 | |
| MgCl2 | 25 mmol·L-1 | BSA | 0.1% | ||
| Lysis buffer | HEPES-KOH, pH7.2 | 50 mmol·L-1 | EDTA | 2 mmol·L-1 | |
| NaCl | 150 mmol·L-1 | Wash buffer | HEPES-KOH, pH7.5 | 20 mmol·L-1 | |
| EDTA | 1 mmol·L-1 | NaCl | 150 mmol·L-1 | ||
| TritonX-100 | 1% | Spermidine | 0.5 mmol·L-1 | ||
| Glycerol | 10% | BSA | 0.1% | ||
| β-mercaptoethanol | 5 mmol·L-1 | Pepstatin A | 1 μg·mL-1 | ||
| Pepstatin A | 1 μg·mL-1 | Aprotinin | 1 μg·mL-1 | ||
| Aprotinin | 1 μg·mL-1 | PMSF | 1 mmol·L-1 | ||
| PMSF | 1 mmol·L-1 | TE buffer | Tris-HCl, pH8.0 | 1 mmol·L-1 | |
| HBB buffer | Tris-HCl, pH7.5 | 25 mmol·L-1 | EDTA | 0.1 mmol·L-1 | |
| Sucrose | 0.44 mol·L-1 | Dissolution buffer | NaCl | 300 mmol·L-1 | |
| MgCl2 | 10 mmol·L-1 | EDTA | 1 mmol·L-1 | ||
| TritonX-100 | 0.1% | ||||
| β-mercaptoethanol | 10 mmol·L-1 |
Table 1 Buffer used in this study
| Buffer | Component | Concentration | Buffer | Component | Concentration |
|---|---|---|---|---|---|
| HEGX buffer | NaCl | 0.8 mol·L-1 | HBC buffer | Tris-HCl, pH7.5 | 20 mmol·L-1 |
| Glycerol | 10% | Sucrose | 352 mmol·L-1 | ||
| TritonX-100 | 0.2% | MgCl2 | 8 mmol·L-1 | ||
| HEPES-KOH, pH7.2 | 20 mmol·L-1 | TritonX-100 | 0.08% | ||
| EDTA | 1 mmol·L-1 | β-mercaptoethanol | 8 mmol·L-1 | ||
| Storage buffer | NaCl | 0.8 mol·L-1 | Glycerol | 20% | |
| Glycerol | 10% | 10× binding buffer | HEPES-KOH, pH8.0 | 20 mmol·L-1 | |
| Tris-HCl, pH7.5 | 50 mmol·L-1 | KCl | 100 mmol·L-1 | ||
| EDTA | 0.2 mmol·L-1 | CaCl2 | 10 mmol·L-1 | ||
| DTT | 2 mmol·L-1 | MnCl2 | 10 mmol·L-1 | ||
| Annealing buffer | Tris-HCl, pH7.0 | 5 mmol·L-1 | Spermidine | 5 mmol·L-1 | |
| EDTA | 0.5 mmol·L-1 | Blocking buffer | HEPES-KOH, pH7.5 | 20 mmol·L-1 | |
| NaCl | 50 mmol·L-1 | NaCl | 150 mmol·L-1 | ||
| 5× Tagmentation buffer | TAPS-NaOH, pH8.5 | 50 mmol·L-1 | Spermidine | 0.5 mmol·L-1 | |
| MgCl2 | 25 mmol·L-1 | BSA | 0.1% | ||
| Lysis buffer | HEPES-KOH, pH7.2 | 50 mmol·L-1 | EDTA | 2 mmol·L-1 | |
| NaCl | 150 mmol·L-1 | Wash buffer | HEPES-KOH, pH7.5 | 20 mmol·L-1 | |
| EDTA | 1 mmol·L-1 | NaCl | 150 mmol·L-1 | ||
| TritonX-100 | 1% | Spermidine | 0.5 mmol·L-1 | ||
| Glycerol | 10% | BSA | 0.1% | ||
| β-mercaptoethanol | 5 mmol·L-1 | Pepstatin A | 1 μg·mL-1 | ||
| Pepstatin A | 1 μg·mL-1 | Aprotinin | 1 μg·mL-1 | ||
| Aprotinin | 1 μg·mL-1 | PMSF | 1 mmol·L-1 | ||
| PMSF | 1 mmol·L-1 | TE buffer | Tris-HCl, pH8.0 | 1 mmol·L-1 | |
| HBB buffer | Tris-HCl, pH7.5 | 25 mmol·L-1 | EDTA | 0.1 mmol·L-1 | |
| Sucrose | 0.44 mol·L-1 | Dissolution buffer | NaCl | 300 mmol·L-1 | |
| MgCl2 | 10 mmol·L-1 | EDTA | 1 mmol·L-1 | ||
| TritonX-100 | 0.1% | ||||
| β-mercaptoethanol | 10 mmol·L-1 |
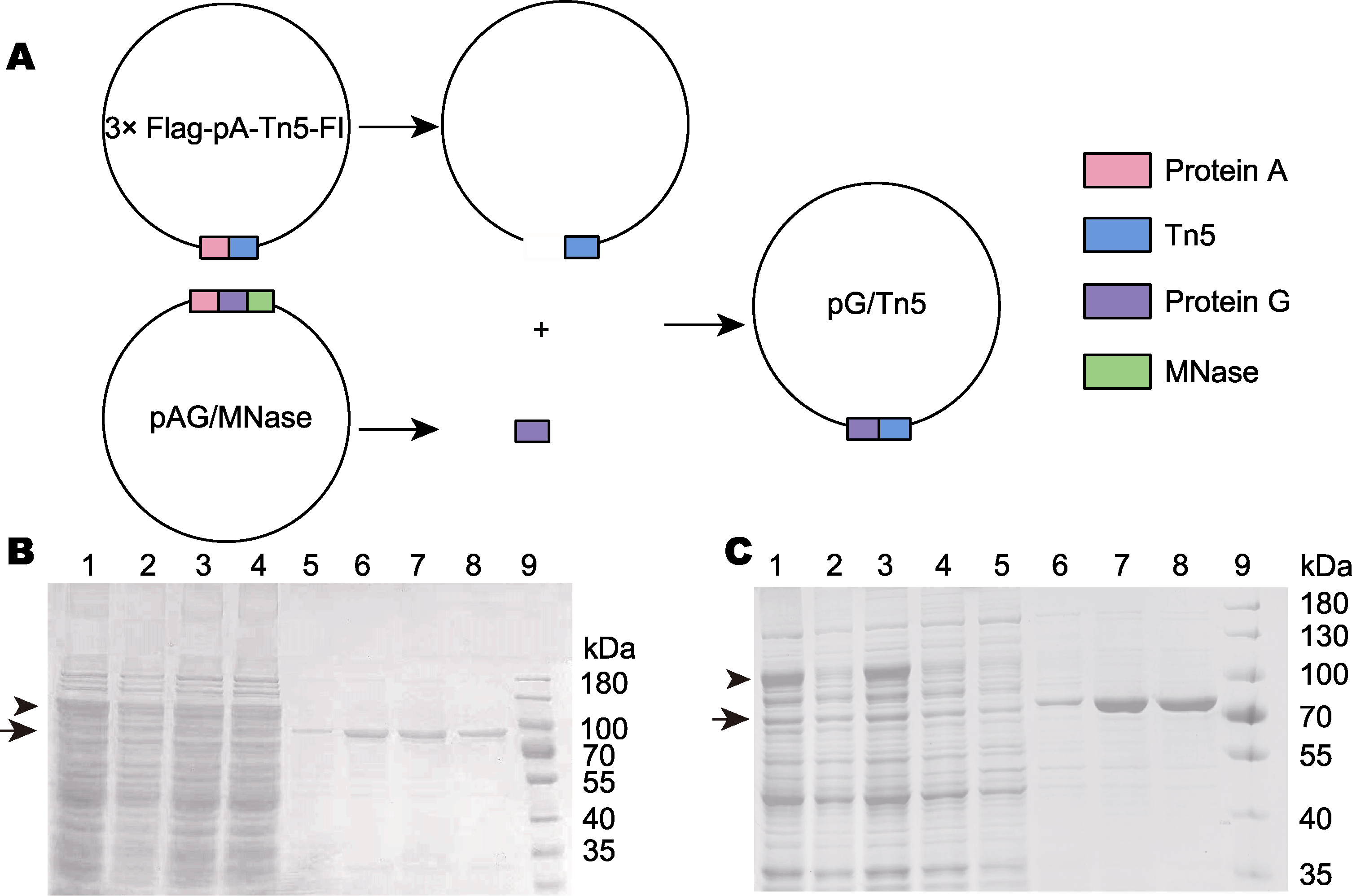
Figure 2 Construction of the pG/Tn5 expression vector and preparation of pA-Tn5 and pG-Tn5 (A) pG/Tn5 was constructed by PCR and homologous recombination; (B) pA-Tn5 was affinity purified by the chitin-binding domain (lanes 1-4 are the protein mixture before purification, represents the protein before purification; lanes 5-8 are pA-Tn5 with high purity after elution, represents the purified protein; lane 9: Protein molecular weight marker); (C) pG-Tn5 is affinity purified by chitin-binding domain (lanes 1-5 are the protein mixture before purification, represents the protein before purification; lanes 6-8 are pG-Tn5 with high purity after elution, represents the purified protein; lane 9: Protein molecular weight marker)
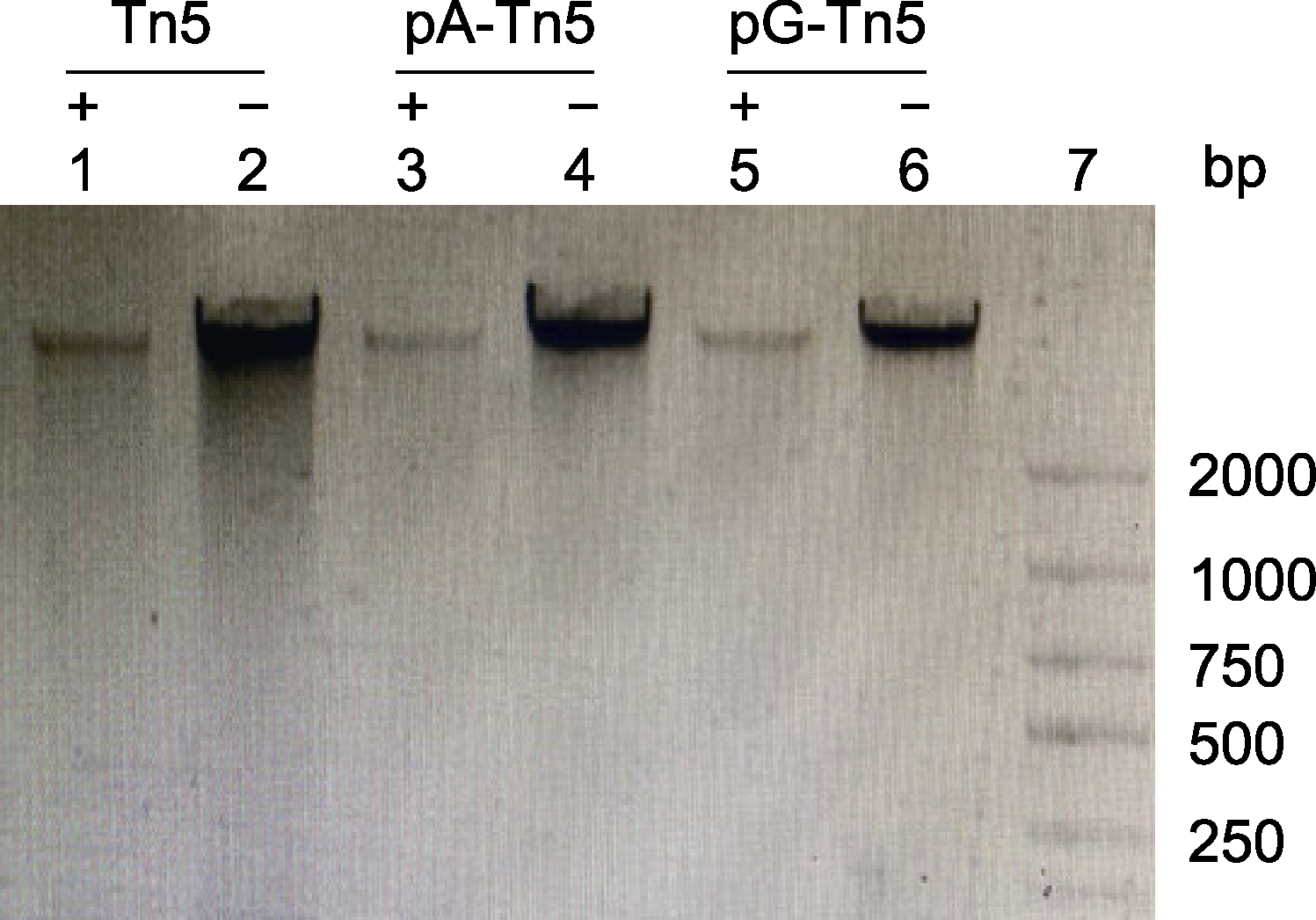
Figure 2 Evaluation of the activities of pA-Tn5 and pG-Tn5 Lane 1: Soybean genomic DNA+Tn5; Lane 3: Soybean genomic DNA+pA-Tn5; Lane 5: Soybean genomic DNA+pG- Tn5; Lanes 2, 4 and 6: Soybean genomic DNA without enzyme; Lane 7: Marker
| Group | Enzyme type | Antibody | Dosage of Tn5 (μg) | Dosage of plant material (μL) |
|---|---|---|---|---|
| 1 | pA-Tn5 | Ser5P Mouse IgG1 | 0.2 | 150 |
| 2 | pA-Tn5 | Ser5P Mouse IgG1 | 1 | 150 |
| 3 | pG-Tn5 | Ser5P Mouse IgG1 | 0.2 | 150 |
| 4 | pG-Tn5 | Ser5P Mouse IgG1 | 1 | 150 |
| 5 | pG-Tn5 | IgG | 0.2 | 150 |
| 6 | pG-Tn5 | IgG | 1 | 150 |
| 7 | pA-Tn5 | Ser5P Rabbit IgG | 0.5 | 30 |
| 8 | pA-Tn5 | Ser5P Rabbit IgG | 1 | 150 |
| 9 | pG-Tn5 | Ser5P Rabbit IgG | 0.5 | 30 |
| 10 | pG-Tn5 | Ser5P Rabbit IgG | 1 | 150 |
| 11 | pA-Tn5 | IgG | 1 | 150 |
| 12 | pG-Tn5 | IgG | 1 | 150 |
Table 2 Experimental group design
| Group | Enzyme type | Antibody | Dosage of Tn5 (μg) | Dosage of plant material (μL) |
|---|---|---|---|---|
| 1 | pA-Tn5 | Ser5P Mouse IgG1 | 0.2 | 150 |
| 2 | pA-Tn5 | Ser5P Mouse IgG1 | 1 | 150 |
| 3 | pG-Tn5 | Ser5P Mouse IgG1 | 0.2 | 150 |
| 4 | pG-Tn5 | Ser5P Mouse IgG1 | 1 | 150 |
| 5 | pG-Tn5 | IgG | 0.2 | 150 |
| 6 | pG-Tn5 | IgG | 1 | 150 |
| 7 | pA-Tn5 | Ser5P Rabbit IgG | 0.5 | 30 |
| 8 | pA-Tn5 | Ser5P Rabbit IgG | 1 | 150 |
| 9 | pG-Tn5 | Ser5P Rabbit IgG | 0.5 | 30 |
| 10 | pG-Tn5 | Ser5P Rabbit IgG | 1 | 150 |
| 11 | pA-Tn5 | IgG | 1 | 150 |
| 12 | pG-Tn5 | IgG | 1 | 150 |
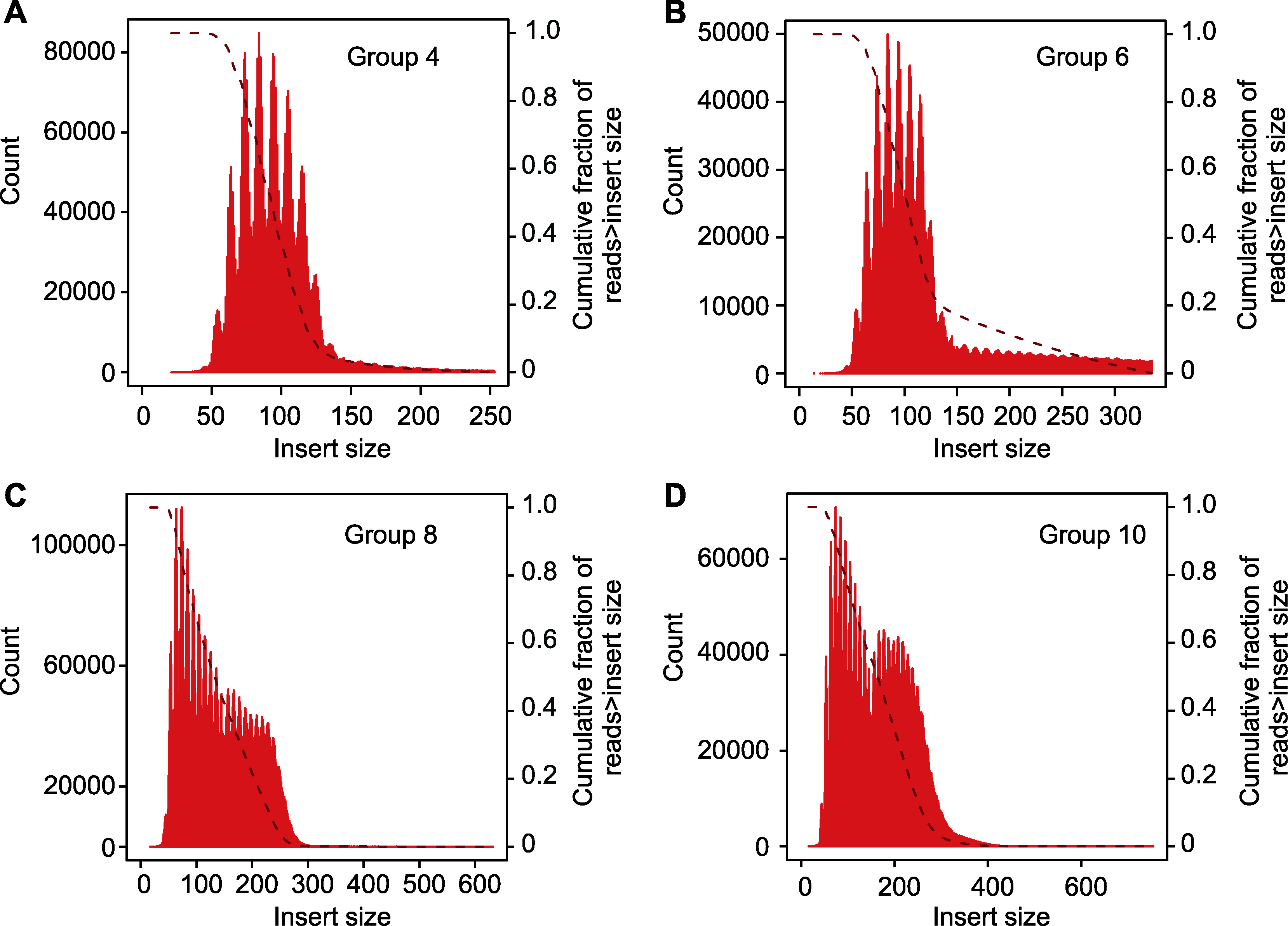
Figure 3 Library insert size (A) Library insert size in experimental group 4; (B) Library insert size in experimental group 6; (C) Library insert size in experimental group 8; (D) Library insert size in experimental group 10. Group 4, 6, 8 and 10 are the same as shown in Table 2.
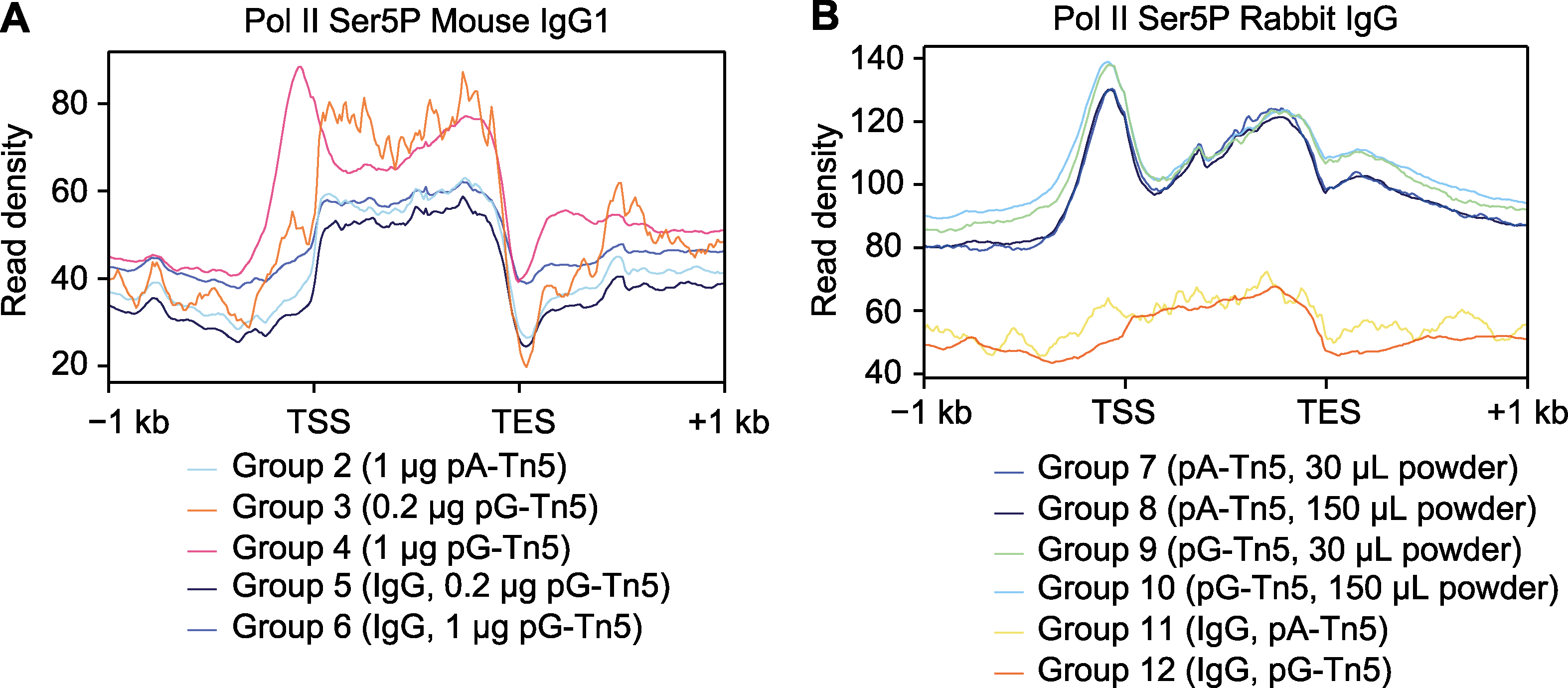
Figure 4 Average distribution of CUT&Tag signals on genes (A) Average distribution of CUT&Tag signals on genes of mouse IgG1 antibody; (B) Average distribution of CUT&Tag signals on genes of rabbit IgG antibody. TSS: Transcription start site; TES: Transcription end site
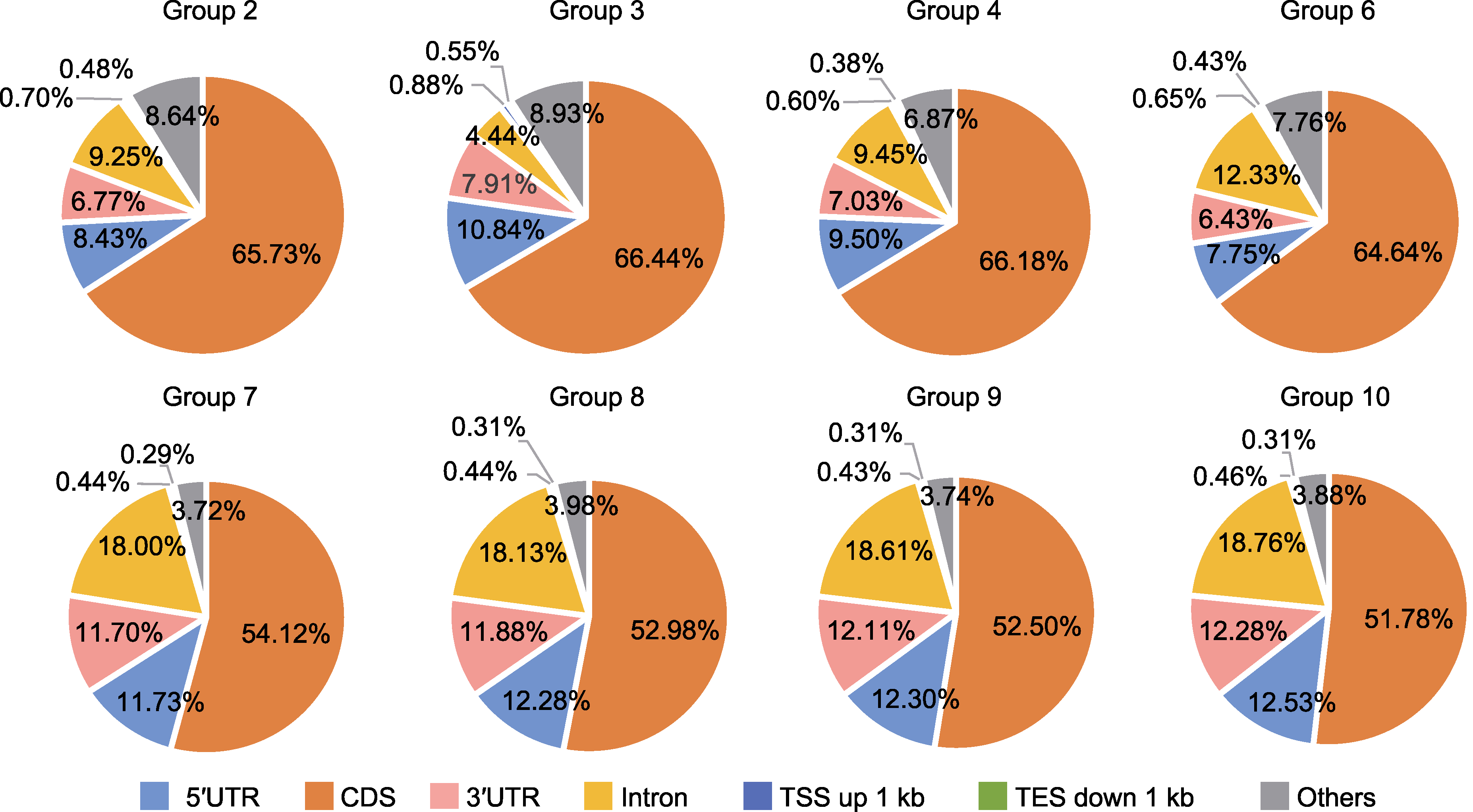
Figure 5 Distribution of CUT&Tag signals on chromosome CDS: Coding sequence; 5′UTR: 5′ untranslated region; 3′UTR: 3′ untranslated region. TSS and TES are the same as shown in Figure 4. Group 2, 3, 4, 6, 7, 8, 9 and 10 are the same as shown in Table 2.
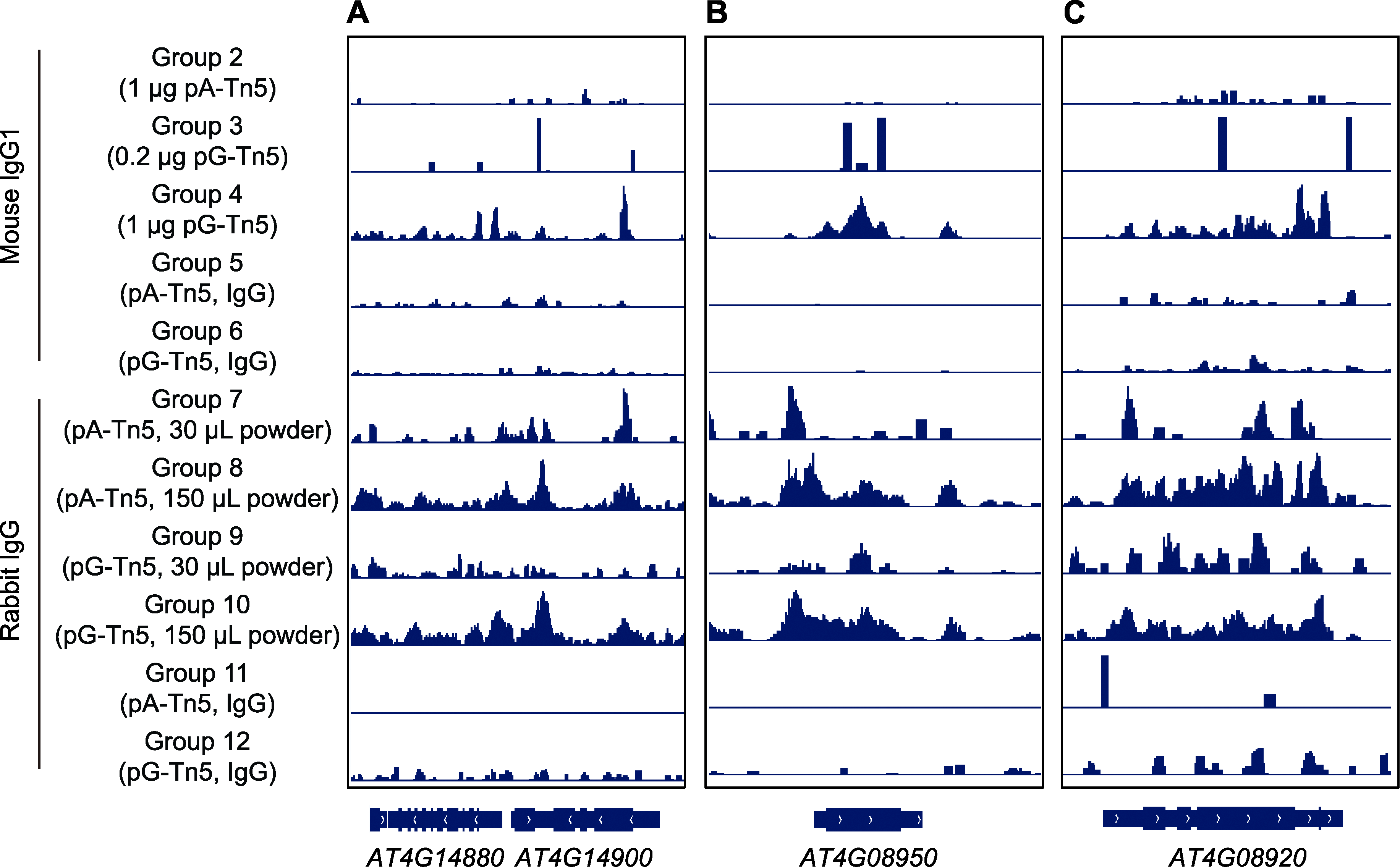
Figure 6 Distribution of CUT&Tag signals on individual genes by IGV (A) CUT&Tag signal of gene AT4G14880 to AT4G14900; (B) CUT&Tag signals of phosphorus responsive protein gene EXO; (C) CUT&Tag signals of blue light receptor gene CRY1. From top to bottom, it corresponds to the experimental data of experimental groups 2-12.
| [1] |
陈威, 杨颖增, 陈锋, 周文冠, 舒凯 (2019). 表观遗传修饰介导的植物胁迫记忆. 植物学报 54, 779-785.
DOI |
| [2] |
杜康兮, 沈文辉, 董爱武 (2018). 表观遗传调控植物响应非生物胁迫的研究进展. 植物学报 53, 581-593.
DOI |
| [3] |
王泓力, 焦雨铃 (2020). 染色质免疫共沉淀实验方法. 植物学报 55, 475-480.
DOI |
| [4] |
杨同文, 李成伟 (2014). 植物叶片衰老的表观遗传调控. 植物学报 49, 729-737.
DOI |
| [5] |
Brahma S, Henikoff S (2019). RSC-associated subnucleosomes define MNase-sensitive promoters in yeast. Mol Cell 73, 238-249.
DOI PMID |
| [6] |
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213-1218.
DOI PMID |
| [7] |
Dancette OP, Taboureau JL, Tournier E, Charcosset C, Blond P (1999). Purification of immunoglobulins G by protein A/G affinity membrane chromatography. J Chromatogr B: Biomed Sci Appl 723, 61-68.
DOI URL |
| [8] | Di L, Fu Y, Sun YS, Li J, Liu L, Yao JC, Wang GB, Wu YL, Lao KQ, Lee RW, Zheng GH, Xu J, Oh J, Wang D, Xie XS, Huang YY, Wang JB (2020). RNA sequencing by direct tagmentation of RNA/DNA hybrids. Proc Natl Acad Sci USA 117, 2886-2893. |
| [9] |
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21.
DOI PMID |
| [10] |
Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S (2019). CUT& Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10, 1930.
DOI PMID |
| [11] |
Kharchenko PV, Tolstorukov MY, Park PJ (2008). Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26, 1351-1359.
DOI PMID |
| [12] |
Lindmark R, Thorén-Tolling K, Sjöquist J (1983). Binding of immunoglobulins to protein A and immunoglobulin levels in mammalian sera. J Immunol Methods 62, 1-13.
PMID |
| [13] |
Lovell S, Goryshin IY, Reznikoff WR, Rayment I (2002). Two-metal active site binding of a Tn5 transposase synaptic complex. Nat Struct Biol 9, 278-281.
PMID |
| [14] | Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17, 10-12. |
| [15] | Meers MP, Bryson TD, Henikoff JG, Henikoff S (2019). Improved CUT&RUN chromatin profiling tools. eLife 8, e46314. |
| [16] | Ouyang WZ, Zhang XW, Peng Y, Zhang Q, Cao ZL, Li GL, Li XW (2021). Rapid and low-input profiling of histone marks in plants using nucleus CUT&Tag. Front Plant Sci 12, 734679. |
| [17] |
Picelli S, Björklund ÅK, Reinius B, Sagasser S, Winberg G, Sandberg R (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24, 2033-2040.
DOI PMID |
| [18] | Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T (2016). DeepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160-W165. |
| [19] |
Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT (2009). ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240-248.
DOI PMID |
| [20] | Skene PJ, Henikoff S (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856. |
| [21] |
Tao XY, Feng SL, Zhao T, Guan XY (2020). Efficient chromatin profiling of H3K4me3 modification in cotton using CUT&Tag. Plant Methods 16, 120.
DOI |
| [22] |
Thakur J, Henikoff S (2018). Unexpected conformational variations of the human centromeric chromatin complex. Genes Dev 32, 20-25.
DOI URL |
| [23] |
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178-192.
DOI PMID |
| [24] |
Wang LG, Wang SQ, Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184-2185.
DOI PMID |
| [25] |
Zheng XY, Gehring M (2019). Low-input chromatin profiling in Arabidopsis endosperm using CUT&RUN. Plant Reprod 32, 63-75.
DOI |
| [1] | DENG Wen-Jie, WU Hua-Zheng, LI Tian-Xiang, ZHOU Li-Na, HU Ren-Yong, JIN Xin-Jie, ZHANG Yong-Pu, ZHANG Yong-Hua, LIU Jin-Liang. Main vegetation types and characteristics in Dongtou National Marine Park, Zhejiang, China [J]. Chin J Plant Ecol, 2024, 48(2): 254-268. |
| [2] | Min Kang. Wallace’s anthropological thought and its contemporary value [J]. Biodiv Sci, 2023, 31(12): 23304-. |
| [3] | Hongli Wang,Yuling Jiao. Protocols for Chromatin Immunoprecipitation [J]. Chinese Bulletin of Botany, 2020, 55(4): 475-480. |
| [4] | Qianqian Li, Yang Jiao, Jingyang Yu, Qiuli Li. Analysis of Differentially Expressed Genes in Suaeda liaotungensis NAC4 (SlNAC4)-transgenic Arabidopsis [J]. Chinese Bulletin of Botany, 2016, 51(6): 764-773. |
| [5] | Xuemei Zhang, Xufang Han, Liwei Liu, Aichun Xu. Influencing factors of the nested distribution of butterfly assemblages in the Zhoushan Archipelago, China [J]. Biodiv Sci, 2016, 24(3): 321-331. |
| [6] | Minghong Wang, Lai Ma, Xiaojiang Zheng, Yibing Hu. Plant Microfluidic Chip, an Integrated High-throughput Platform for Real-time Analysis of Plant Growth and Development [J]. Chinese Bulletin of Botany, 2015, 50(5): 637-. |
| [7] | Naicheng Li, Xiaoshou Liu, Zhaodong Xu, Rui Zhao, Honghua Shi. Biodiversity of macrofauna in the southern waters of Miaodao Archipelago, China [J]. Biodiv Sci, 2015, 23(1): 41-49. |
| [8] | Xin Sun,Ying Gao,Yunfeng Yang. Recent advancement in microbial environmental research using metagenomics tools [J]. Biodiv Sci, 2013, 21(4): 393-400. |
| [9] | Zhenhua Wang, Shouyu Zhang, Qingman Chen, Qiang Xu, Kai Wang. Fish community ecology in rocky reef habitat of Ma’an Archipelago. I. Species composition and diversity [J]. Biodiv Sci, 2012, 20(1): 41-50. |
| [10] | Bin Liang, Shuihua Chen, Zhongde Wang . Nest selection of Chinese egret (Egretta eulophotes) in Wuzhishan Archi-pelago, Zhejiang [J]. Biodiv Sci, 2007, 15(1): 92-96. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||