

Chinese Bulletin of Botany ›› 2020, Vol. 55 ›› Issue (1): 1-4.DOI: 10.11983/CBB19240 cstr: 32102.14.CBB19240
• COMMENTARIES • Next Articles
Jiali Tang,Jie Qiu,Xuehui Huang( )
)
Received:2019-12-15
Accepted:2019-12-17
Online:2020-01-01
Published:2019-12-20
Contact:
Xuehui Huang
Jiali Tang,Jie Qiu,Xuehui Huang. The Development of Genomics Technologies Drives New Progress in Horticultural Plant Research[J]. Chinese Bulletin of Botany, 2020, 55(1): 1-4.
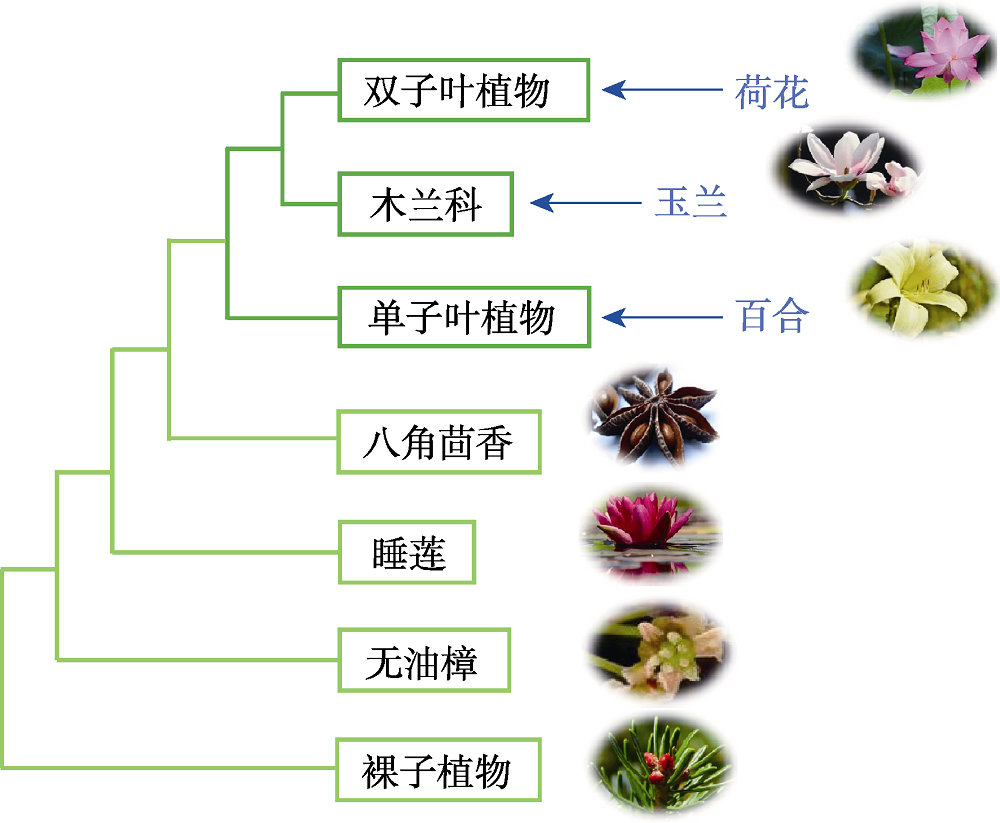
Figure 1 Plant phylogeny diagram According to plant phylogeny, water lilies belong to primitive angiosperms, while lotuses belong to typical dicotyledons.
| [1] | Chen F, Liu X, Yu CW, Chen YC, Tang HB, Zhang LS (2017). Water lilies as emerging models for Darwin’s abominable mystery. Hortic Res 4, 17051. |
| [2] | Chen LY, VanBuren R, Paris M, Zhou HY, Zhang XT, Wai CM, Yan HS, Chen S, Alonge M, Ramakrishnan S, Liao ZY, Liu J, Lin JS, Yue JJ, Fatima M, Lin ZC, Zhang JS, Huang LX, Wang H, Hwa TY, Kao SM, Choi AEY, Sharma A, Song J, Wang LL, Yim WC, Cushman JC, Paull RE, Matsumoto T, Qin Y, Wu QS, Wang JP, Yu QY, Wu J, Zhang SL, Boches P, Tung CW, Wang ML, D’Eeckenbrugge GC, Sanewski GM, Purugganan MD, Schatz MC, Bennetzen JL, Lexer C, Ming R (2019). The bracteatus pineapple genome and domestication of clonally propagated crops. Nat Genet 51, 1549-1558. |
| [3] | Guo SG, Zhao SJ, Sun HH, Wang X, Wu S, Lin T, Ren Y, Gao L, Deng Y, Zhang J, Lu XQ, Zhang HY, Shang JL, Gong GY, Wen CL, He N, Tian SW, Li MY, Liu JP, Wang YP, Zhu YC, Jarrets R, Levi A, Zhang XP, Huang SW, Fei ZJ, Liu WG, Xu Y (2019). Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat Genet 51, 1616-1623. |
| [4] | Liao NQ, Hu ZY, Li YY, Hao JF, Chen SN, Xue Q, Ma YY, Zhang KJ, Mahmoud A, Ali A, Malangisha GK, Lyu XL, Yang JH, Zhang MF (2020). Ethylene-responsive factor 4 is associated with the desirable rind hardness trait conferring cracking resistance in fresh fruits of watermelon. Plant Biol J 18, 1066-1077. |
| [5] | Ming R, VanBuren R, Wai CM, Tang HB, Schatz MC, Bowers JE, Lyons E, Wang ML, Chen J, Biggers E, Zhang JS, Huang LX, Zhang LM, Miao WJ, Zhang J, Ye ZY, Miao CY, Lin ZC, Wang H, Zhou HY, Yim WC, Priest HD, Zheng CF, Woodhouse M, Edger PP, Guyot R, Guo HB, Guo H, Zheng GY, Singh R, Sharma A, Min XJ, Zheng Y, Lee H, Gurtowski J, Sedlazeck FJ, Harkess A, McKain MR, Liao ZY, Fang JP, Liu J, Zhang XD, Zhang Q, Hu WC, Qin Y, Wang K, Chen LY, Shirley N, Lin YR, Liu LY, Hernandez AG, Wright CL, Bulone V, Tuskan GA, Heath K, Zee F, Moore PH, Sunkar R, Leebens-Mack JH, Mockler T, Bennetzen JL, Freeling M, Sankoff D, Paterson AH, Zhu XG, Yang XH, Smith JAC, Cushman JC, Paull RE, Yu QY (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47, 1435-1442. |
| [6] | The French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463-467. |
| [7] | Zhang LS, Chen F, Zhang XT, Li Z, Zhao YY, Rolf L, Chang XJ, Dong W, Simon YWH, Liu X, Song AX, Chen JH, Guo WL, Wang ZJ, Zhuang YY, Wang HF, Chen XQ, Hu J, Liu YH, Qin Y, Wang K, Dong SS, Liu Y, Zhang SZ, Yu XX, Wu Q, Wang LS, Yan XQ, Jiao YN, Kong HZ, Zhou XF, Yu CW, Chen YC, Li F, Wang JH, Chen W, Chen XL, Jia QD, Zhang C, Jiang YF, Zhang WB, Liu GH, Fu JY, Chen F, Ma H, Yves VP, Tang HB (2019). The water lily genome and the early evolution of flowering plants. Nature 577, 79-84. |
| [8] | Zhao GW, Lian Q, Zhang ZH, Fu QS, He YH, Ma S, Ruggieri V, Monforte AJ, Wang PY, Julca I, Wang HS, Liu JP, Xu Y, Wang RZ, Ji JB, Xu ZH, Kong WH, Zhong Y, Shang JL, Pereira L, Argyris J, Zhang J, Mayobre C, Pujol M, Oren E, Out D, Wang JM, Sun DX, Zhao SJ, Zhu YC, Li N, Katzir N, Gur A, Dogimont C, Schaefer H, Fan W, Bendahmane A, Fei Z, Pitrat M, Gabaldon T, Lin T, Garcia-Mas J, Xu YY, Huang SW (2019). A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat Genet 51, 1607-1615. |
| [1] | Chuanyong Wang, Dian Zhuang, Zhengda Song, Henghua Zhai, Naiwei Li, Fan Zhang. Structural and Comparative Analysis of the Complete Chloroplast Genome and Phylogenetic Inference of the Aronia melanocarpa [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [2] |
Juan Cui, Xiaoyu Yu, Yuejiao Yu, Chengwei Liang, Jian Sun, Wenfu Chen.
Analysis of Texture Factors and Genetic Basis Influencing the Differences in Eating Quality between Northeast China and Japanese Japonica Rice [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [3] | Xiaolin Chu, Quanguo Zhang. A review of experimental evidence for the evolutionary speed hypothesis [J]. Biodiv Sci, 2025, 33(4): 25019-. |
| [4] | Bailong Zhao, Yeliang Li, Yufei Wang, Bin Sun. New Leaf Classification of Mahonia (Berberidaceae) [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [5] | Zhang Ruli, Li Dezhu, Zhang Yuxiao. Population Genetic Structure and Climate Adaptation Analysis of Brachystachyum densiflorum [J]. Chinese Bulletin of Botany, 2025, 60(3): 407-424. |
| [6] | Linfeng Xia, Rui Li, Haizheng Wang, Daling Feng, Chunyang Wang. Research Advances and Prospects in Charophytes Genomics [J]. Chinese Bulletin of Botany, 2025, 60(2): 271-282. |
| [7] | Cao Dong, Li Huanlong, Peng Yang, Wei Cunzheng. Progresses in the study of the relationship between plant genome size and traits [J]. Biodiv Sci, 2025, 33(2): 24192-. |
| [8] | Lin Zhen, Xiang Jiabao, Cai Hejiayi, Gao Bei, Yang Jintao, Li Junyi, Zhou Qingsong, Huang Xiaolei, Deng Jun. Mitochondrial genomic data for seven Hemipteran species [J]. Biodiv Sci, 2025, 33(2): 24434-. |
| [9] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [10] | Yajun Sun. What do higher or lower organisms mean—Clarify the meaning and validity of the biological ladder implied by On the Origin of Species [J]. Biodiv Sci, 2025, 33(1): 24394-. |
| [11] | Xiangtan Yao, Xinyi Zhang, Yang Chen, Ye Yuan, Wangda Cheng, Tianrui Wang, Yingxiong Qiu. Genomic resequencing reveals the genetic diversity of the cultivated water caltrop, and the origin and domestication of ‘Nanhuling’ [J]. Biodiv Sci, 2024, 32(9): 24212-. |
| [12] | Nan Chen, Quan-Guo Zhang. The experimental evolution approach [J]. Biodiv Sci, 2024, 32(9): 24171-. |
| [13] | Qiang Zhang, Zhenyu Zhao, Pinghua Li. Research Progress of Gene Editing Technology in Maize [J]. Chinese Bulletin of Botany, 2024, 59(6): 978-998. |
| [14] | Hua He, Dunyan Tan, Xiaochen Yang. Cryptic dioecy in angiosperms: Diversity, phylogeny and evolutionary significance [J]. Biodiv Sci, 2024, 32(6): 24149-. |
| [15] | Yuan Li, Kaijian Fan, Tai An, Cong Li, Junxia Jiang, Hao Niu, Weiwei Zeng, Yanfang Heng, Hu Li, Junjie Fu, Huihui Li, Liang Li. Study on Multi-environment Genome-wide Prediction of Inbred Agronomic Traits in Maize Natural Populations [J]. Chinese Bulletin of Botany, 2024, 59(6): 1041-1053. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||