

Chinese Bulletin of Botany ›› 2015, Vol. 50 ›› Issue (5): 549-564.DOI: 10.11983/CBB15164 cstr: 32102.14.CBB15164
Previous Articles Next Articles
Mian Liu1,2, Caifei Zhang1,2*, Chien-Hsun Huang1,2, Hong Ma1,2*
Received:2015-08-15
Accepted:2015-09-05
Online:2015-09-01
Published:2015-10-09
Contact:
Zhang Caifei,Ma Hong
About author:? These authors contributed equally to this paper
Mian Liu, Caifei Zhang, Chien-Hsun Huang, Hong Ma. Phylogenetic Reconstruction of Tribal Relationships in Asteroi- deae (Asteraceae) with Low-copy Nuclear Genes[J]. Chinese Bulletin of Botany, 2015, 50(5): 549-564.
| Tribe | Species | Locality | Accession No. | Voucher |
|---|---|---|---|---|
| Anthemideae | Achillea millefolium | Campus of Penn State University, USA | - | Zhang Ning, B20AM |
| Anthemideae | Artemisia chamaemelifolia | NCBI | SRX096997 | - |
| Anthemideae | Tanacetum parthenium | NCBI | ERX337163 | - |
| Astereae | Symphyotrichum subulatum | Chenshan Botanical Garden, Shanghai | - | Liu Mian, CSLM02 |
| Astereae | Eurybia divaricata | Campus of Penn State University, USA | - | Ma Hong, HM213 |
| Astereae | Solidago gigantea | Penn State College, USA | - | Ma Hong, HM157 |
| Athroismeae | Centipeda minima | Beijing Botanical Garden of CAS | - | Gao Tiangang, GTG03 |
| Calenduleae | Osteospermum jucundum | Glasgow, UK | - | Ma Hong, HM366 |
| Calenduleae | Calendula officinalis | Beijing Botanical Garden, Beijing | - | Zhang Caifei, 3395 |
| Cichorieae | Sonchus asper | Campus of Penn State University, USA | - | Zhang Ning, A10SA |
| Cichorieae | Lactuca serriola | NCBI | SRX098217 | - |
| Coreopsideae | Cosmos bipinnatus | Xuancheng, Anhui | - | Liu Mian, AH06 |
| Coreopsideae | Bidens frondosa | Campus of Penn State College, USA | - | Ma Hong, HM208 |
| Eupatorieae | Stevia rebaudiana | NCBI | - | - |
| Eupatorieae | Ageratum conyzoides | Mt. Tianmu, Zhejiang | SRX255224 | Liu Mian, TMS08 |
| Eupatorieae | Mikania micrantha | Indonesia | - | Gao Tiangang, GTG5498 |
| Gnaphalieae | Helichrysum petiolare | NCBI | SRX202783 | - |
| Gnaphalieae | Leontopodium smithianum | Mt. Dongling, Beijing | - | Yang Ji, YJ08 |
| Helenieae | Gaillardia pulchella | Chenshan Botanical Garden, Shanghai | - | Liu Mian, CSLM07 |
| Heliantheae | Helianthus tuberosus | Mt. Tianmu, Zhejiang | - | Liu Mian, TMS15 |
| Heliantheae | Iva annua | NCBI | SRX255215 | - |
| Heliantheae | Echinacea purpurea | NCBI | SRX085117 | - |
| Inuleae | Duhaldea cappa | Campus of Nanjing University, Nanjing | - | Yang Ji, YJLM09 |
| Inuleae | Inula linariifolia | Mt. Zijin, Nanjing | - | Liu Mian, YJLM04 |
| Madieae | Arnica montana | NCBI | SRX096987 | - |
| Millerieae | Sigesbeckia pubescens | Mt. Tianmu, Zhejiang | - | Liu Mian, TMS01 |
| Millerieae | Galinsoga quadriradiata | Mt. Tianmu, Zhejiang | - | Liu Mian, TMS02 |
| Millerieae | Guizotia scabra | NCBI | SRX255223 | - |
| Senecioneae | Euryops pectinatus | Campus of Fudan University, Shanghai | - | Liu Mian, FDU01 |
| Senecioneae | Senecio aethnensis | NCBI | SRX328503 | - |
| Senecioneae | Petasites hybridus | NCBI | SRX096999 | - |
Table 1 Samples used in this study and their information
| Tribe | Species | Locality | Accession No. | Voucher |
|---|---|---|---|---|
| Anthemideae | Achillea millefolium | Campus of Penn State University, USA | - | Zhang Ning, B20AM |
| Anthemideae | Artemisia chamaemelifolia | NCBI | SRX096997 | - |
| Anthemideae | Tanacetum parthenium | NCBI | ERX337163 | - |
| Astereae | Symphyotrichum subulatum | Chenshan Botanical Garden, Shanghai | - | Liu Mian, CSLM02 |
| Astereae | Eurybia divaricata | Campus of Penn State University, USA | - | Ma Hong, HM213 |
| Astereae | Solidago gigantea | Penn State College, USA | - | Ma Hong, HM157 |
| Athroismeae | Centipeda minima | Beijing Botanical Garden of CAS | - | Gao Tiangang, GTG03 |
| Calenduleae | Osteospermum jucundum | Glasgow, UK | - | Ma Hong, HM366 |
| Calenduleae | Calendula officinalis | Beijing Botanical Garden, Beijing | - | Zhang Caifei, 3395 |
| Cichorieae | Sonchus asper | Campus of Penn State University, USA | - | Zhang Ning, A10SA |
| Cichorieae | Lactuca serriola | NCBI | SRX098217 | - |
| Coreopsideae | Cosmos bipinnatus | Xuancheng, Anhui | - | Liu Mian, AH06 |
| Coreopsideae | Bidens frondosa | Campus of Penn State College, USA | - | Ma Hong, HM208 |
| Eupatorieae | Stevia rebaudiana | NCBI | - | - |
| Eupatorieae | Ageratum conyzoides | Mt. Tianmu, Zhejiang | SRX255224 | Liu Mian, TMS08 |
| Eupatorieae | Mikania micrantha | Indonesia | - | Gao Tiangang, GTG5498 |
| Gnaphalieae | Helichrysum petiolare | NCBI | SRX202783 | - |
| Gnaphalieae | Leontopodium smithianum | Mt. Dongling, Beijing | - | Yang Ji, YJ08 |
| Helenieae | Gaillardia pulchella | Chenshan Botanical Garden, Shanghai | - | Liu Mian, CSLM07 |
| Heliantheae | Helianthus tuberosus | Mt. Tianmu, Zhejiang | - | Liu Mian, TMS15 |
| Heliantheae | Iva annua | NCBI | SRX255215 | - |
| Heliantheae | Echinacea purpurea | NCBI | SRX085117 | - |
| Inuleae | Duhaldea cappa | Campus of Nanjing University, Nanjing | - | Yang Ji, YJLM09 |
| Inuleae | Inula linariifolia | Mt. Zijin, Nanjing | - | Liu Mian, YJLM04 |
| Madieae | Arnica montana | NCBI | SRX096987 | - |
| Millerieae | Sigesbeckia pubescens | Mt. Tianmu, Zhejiang | - | Liu Mian, TMS01 |
| Millerieae | Galinsoga quadriradiata | Mt. Tianmu, Zhejiang | - | Liu Mian, TMS02 |
| Millerieae | Guizotia scabra | NCBI | SRX255223 | - |
| Senecioneae | Euryops pectinatus | Campus of Fudan University, Shanghai | - | Liu Mian, FDU01 |
| Senecioneae | Senecio aethnensis | NCBI | SRX328503 | - |
| Senecioneae | Petasites hybridus | NCBI | SRX096999 | - |
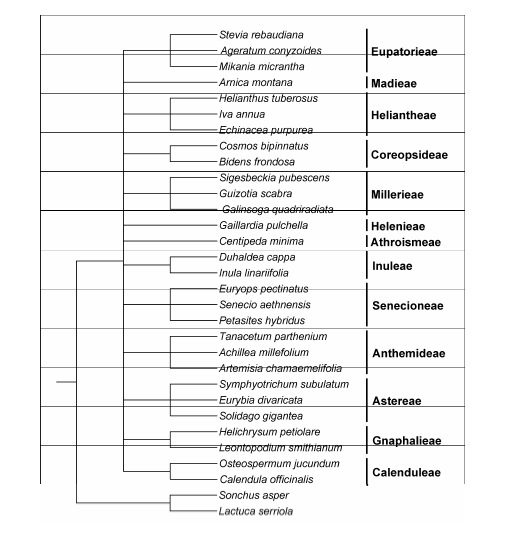
Figure 2 Reference species tree of Asteroideae for selecting orthologous genesIn this tree, the relationships among the tribes, or those between genera of the same tribe, are not specified.
| Unigene | Length (bp) | VS | VS (%) | IS | IS (%) | MS (%) |
|---|---|---|---|---|---|---|
| 433680 | 1 290 | 809 | 62.71 | 637 | 49.38 | 1.67 |
| 433783 | 462 | 293 | 63.42 | 228 | 49.35 | 0.89 |
| 433651 | 1 158 | 656 | 56.65 | 530 | 45.77 | 4.54 |
| 435322 | 1 281 | 716 | 55.89 | 553 | 43.17 | 3.74 |
| 435258 | 1 236 | 707 | 57.20 | 518 | 41.91 | 13.26 |
| 431587 | 864 | 479 | 55.44 | 362 | 41.90 | 2.49 |
| 432845 | 675 | 368 | 54.52 | 276 | 40.89 | 1.81 |
| 431540 | 1 152 | 637 | 55.30 | 470 | 40.80 | 3.73 |
| 432103 | 741 | 394 | 53.17 | 296 | 39.95 | 8.29 |
| 435225 | 1 395 | 765 | 54.84 | 539 | 38.64 | 16.34 |
| 432720 | 1 104 | 575 | 52.08 | 408 | 36.96 | 7.53 |
| 432475 | 828 | 402 | 48.55 | 305 | 36.84 | 4.98 |
| 432118 | 588 | 317 | 53.91 | 215 | 36.56 | 4.30 |
| 435156 | 1 740 | 913 | 52.47 | 634 | 36.44 | 22.01 |
| 435256 | 885 | 436 | 49.27 | 322 | 36.38 | 3.75 |
| 435114 | 1 140 | 547 | 47.98 | 406 | 35.61 | 3.58 |
| 434310 | 993 | 459 | 46.22 | 353 | 35.55 | 5.39 |
| 432389 | 1 902 | 928 | 48.79 | 675 | 35.49 | 5.17 |
| 435299 | 591 | 290 | 49.07 | 209 | 35.36 | 3.59 |
| 432317 | 1 320 | 624 | 47.27 | 463 | 35.08 | 3.20 |
| 435127 | 1 569 | 730 | 46.53 | 550 | 35.05 | 10.70 |
| 435193 | 1 491 | 799 | 53.59 | 522 | 35.01 | 10.09 |
| 435154 | 1 374 | 663 | 48.25 | 478 | 34.79 | 4.13 |
| 431821 | 1 323 | 621 | 46.94 | 459 | 34.69 | 5.00 |
| 433235 | 1 560 | 696 | 44.62 | 538 | 34.49 | 2.20 |
| 434795 | 1 164 | 525 | 45.10 | 394 | 33.85 | 2.46 |
| 435112 | 2 835 | 1332 | 46.98 | 949 | 33.47 | 19.43 |
| 433741 | 777 | 326 | 41.96 | 259 | 33.33 | 3.40 |
| 432132 | 807 | 362 | 44.86 | 267 | 33.09 | 3.22 |
| 433969 | 684 | 303 | 44.30 | 225 | 32.89 | 0.22 |
| 433433 | 1 362 | 557 | 40.90 | 441 | 32.38 | 0.23 |
| 435159 | 558 | 244 | 43.73 | 178 | 31.90 | 4.09 |
| 433560 | 951 | 425 | 44.69 | 303 | 31.86 | 2.42 |
| 432107 | 1 188 | 473 | 39.81 | 376 | 31.65 | 1.30 |
| 432280 | 1 209 | 506 | 41.85 | 381 | 31.51 | 2.13 |
| 433255 | 795 | 318 | 40.00 | 249 | 31.32 | 1.41 |
| 435277 | 1 359 | 552 | 40.62 | 419 | 30.83 | 2.44 |
| 435118 | 876 | 388 | 44.29 | 268 | 30.59 | 2.46 |
| 433710 | 969 | 414 | 42.72 | 294 | 30.34 | 1.81 |
| 432233 | 1 059 | 455 | 42.97 | 319 | 30.12 | 3.78 |
| 432926 | 1 014 | 442 | 43.59 | 299 | 29.49 | 5.14 |
| 433740 | 726 | 285 | 39.26 | 212 | 29.20 | 0.95 |
| 433176 | 789 | 329 | 41.70 | 226 | 28.64 | 1.08 |
| 434314 | 942 | 436 | 46.28 | 269 | 28.56 | 3.59 |
| 435311 | 444 | 165 | 37.16 | 125 | 28.15 | 0.20 |
| 434176 | 1 296 | 482 | 37.19 | 351 | 27.08 | 3.19 |
| 433486 | 741 | 292 | 39.41 | 194 | 26.18 | 0.38 |
| Average | 1 090 | 520 | 47.53 | 382 | 34.95 | 4.63 |
Table 2 The systematic information of 47 unigene matrices, respectively
| Unigene | Length (bp) | VS | VS (%) | IS | IS (%) | MS (%) |
|---|---|---|---|---|---|---|
| 433680 | 1 290 | 809 | 62.71 | 637 | 49.38 | 1.67 |
| 433783 | 462 | 293 | 63.42 | 228 | 49.35 | 0.89 |
| 433651 | 1 158 | 656 | 56.65 | 530 | 45.77 | 4.54 |
| 435322 | 1 281 | 716 | 55.89 | 553 | 43.17 | 3.74 |
| 435258 | 1 236 | 707 | 57.20 | 518 | 41.91 | 13.26 |
| 431587 | 864 | 479 | 55.44 | 362 | 41.90 | 2.49 |
| 432845 | 675 | 368 | 54.52 | 276 | 40.89 | 1.81 |
| 431540 | 1 152 | 637 | 55.30 | 470 | 40.80 | 3.73 |
| 432103 | 741 | 394 | 53.17 | 296 | 39.95 | 8.29 |
| 435225 | 1 395 | 765 | 54.84 | 539 | 38.64 | 16.34 |
| 432720 | 1 104 | 575 | 52.08 | 408 | 36.96 | 7.53 |
| 432475 | 828 | 402 | 48.55 | 305 | 36.84 | 4.98 |
| 432118 | 588 | 317 | 53.91 | 215 | 36.56 | 4.30 |
| 435156 | 1 740 | 913 | 52.47 | 634 | 36.44 | 22.01 |
| 435256 | 885 | 436 | 49.27 | 322 | 36.38 | 3.75 |
| 435114 | 1 140 | 547 | 47.98 | 406 | 35.61 | 3.58 |
| 434310 | 993 | 459 | 46.22 | 353 | 35.55 | 5.39 |
| 432389 | 1 902 | 928 | 48.79 | 675 | 35.49 | 5.17 |
| 435299 | 591 | 290 | 49.07 | 209 | 35.36 | 3.59 |
| 432317 | 1 320 | 624 | 47.27 | 463 | 35.08 | 3.20 |
| 435127 | 1 569 | 730 | 46.53 | 550 | 35.05 | 10.70 |
| 435193 | 1 491 | 799 | 53.59 | 522 | 35.01 | 10.09 |
| 435154 | 1 374 | 663 | 48.25 | 478 | 34.79 | 4.13 |
| 431821 | 1 323 | 621 | 46.94 | 459 | 34.69 | 5.00 |
| 433235 | 1 560 | 696 | 44.62 | 538 | 34.49 | 2.20 |
| 434795 | 1 164 | 525 | 45.10 | 394 | 33.85 | 2.46 |
| 435112 | 2 835 | 1332 | 46.98 | 949 | 33.47 | 19.43 |
| 433741 | 777 | 326 | 41.96 | 259 | 33.33 | 3.40 |
| 432132 | 807 | 362 | 44.86 | 267 | 33.09 | 3.22 |
| 433969 | 684 | 303 | 44.30 | 225 | 32.89 | 0.22 |
| 433433 | 1 362 | 557 | 40.90 | 441 | 32.38 | 0.23 |
| 435159 | 558 | 244 | 43.73 | 178 | 31.90 | 4.09 |
| 433560 | 951 | 425 | 44.69 | 303 | 31.86 | 2.42 |
| 432107 | 1 188 | 473 | 39.81 | 376 | 31.65 | 1.30 |
| 432280 | 1 209 | 506 | 41.85 | 381 | 31.51 | 2.13 |
| 433255 | 795 | 318 | 40.00 | 249 | 31.32 | 1.41 |
| 435277 | 1 359 | 552 | 40.62 | 419 | 30.83 | 2.44 |
| 435118 | 876 | 388 | 44.29 | 268 | 30.59 | 2.46 |
| 433710 | 969 | 414 | 42.72 | 294 | 30.34 | 1.81 |
| 432233 | 1 059 | 455 | 42.97 | 319 | 30.12 | 3.78 |
| 432926 | 1 014 | 442 | 43.59 | 299 | 29.49 | 5.14 |
| 433740 | 726 | 285 | 39.26 | 212 | 29.20 | 0.95 |
| 433176 | 789 | 329 | 41.70 | 226 | 28.64 | 1.08 |
| 434314 | 942 | 436 | 46.28 | 269 | 28.56 | 3.59 |
| 435311 | 444 | 165 | 37.16 | 125 | 28.15 | 0.20 |
| 434176 | 1 296 | 482 | 37.19 | 351 | 27.08 | 3.19 |
| 433486 | 741 | 292 | 39.41 | 194 | 26.18 | 0.38 |
| Average | 1 090 | 520 | 47.53 | 382 | 34.95 | 4.63 |

Figure 3 A maximum likelihood tree using 47 nuclear genes for 29 Asteroideae species The numbers on each node are support values of ML and Bayesian analyses, respectively. Asterisks stand for bootstrap of 100% or posterior probability of 1.0.
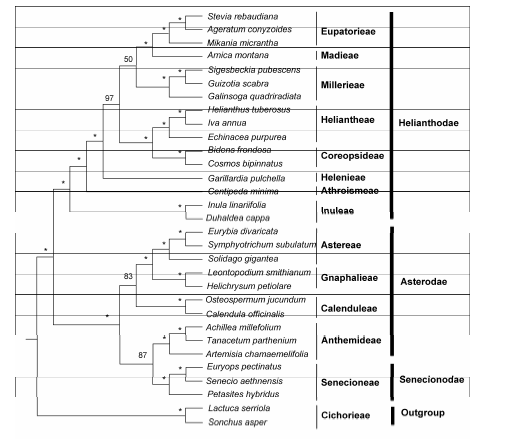
Figure 4 Coalescent-based species tree of Asteroideae estimated from 47 gene treesThe numbers on each node are bootstrap support values. Asterisks stand for bootstrap of 100%.
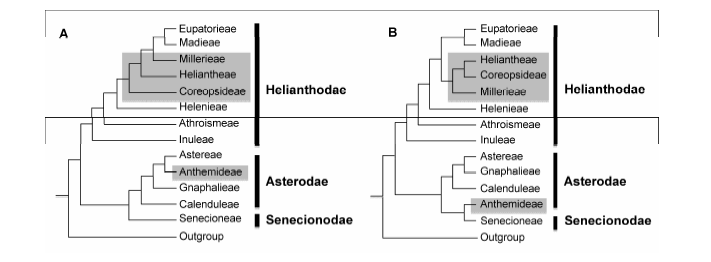
Figure 5 Comparison of Intertribal phylogenies of Asteroideae based on different data sets(A) Cladogram based on chloroplast genes (revised from Panero and Funk (2002, 2008) and Panero et al. (2014)); (B) Cladogram based on concatenated alignment of 47 nuclear genes used by present work. For all clades, bootstrap values of MP or ML analyses are all ≥70%. The shaded areas show obvious incongruences on both trees.
| 1 | 刘勉 (2015). 用低拷贝核基因重建菊科植物的系统发育关系. 硕士论文. 上海: 复旦大学. |
| 2 | Baldwin BG (2009). Heliantheae alliance. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 689-711. |
| 3 | Baldwin BG, Wessa BL, Panero JL (2002). Nuclear rDNA evidence for major lineages of helenioid Heliantheae (Com- positae).Syst Bot 27, 161-198. |
| 4 | Bayer RJ, Starr JR (1998). Tribal phylogeny of the Asteraceae based on two non-coding chloroplast sequ- ences, the trnL intron and trnL/trnF intergenic spacer.Ann Mo Bot Gard 85, 242-256. |
| 5 | Bremer K (1987). Tribal interrelationships of the Aster- aceae.Cladistics 3, 210-253. |
| 6 | Bremer K, Anderberg AA (1994). Asteraceae: Cladistics & Classification. Portland: Timber Press. |
| 7 | Calabria LM, Emerenciano VP, Ferreira MJ, SCotti MT, Mabry TJ (2007). A phylogenetic analysis of tribes of the Asteraceae based on phytochemical data. Nat Prod Com- mun 2, 277-285. |
| 8 | Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses.Bioinformatics 25, 1972-1973. |
| 9 | Corriveau JL, Coleman AW (1988). Rapid screening me- thod to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species.Am J Bot 75, 1443-1458. |
| 10 | Darriba D, Taboada GL, Doallo R, Posada D (2012). jModelTest 2: more models, new heuristics and parallel computing.Nat Methods 9, 772-772. |
| 11 | Ebersberger I, Strauss S, von Haeseler A (2009). Ha- MStR: profile hidden markov model based search for orthologs in ESTs.BMC Evol Biol 9, 157. |
| 12 | Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput.Nucleic Acids Res 32, 1792-1797. |
| 13 | Feliner GN, Rosselló JA (2007). Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants.Mol Phylogenet Evol 44, 911-919. |
| 14 | Funk VA, Susanna A, Stuessy TF, Robinson H (2009). Classification of compositae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 171-192. |
| 15 | Goertzen LR, Cannone JJ, Gutell RR, Jansen RK (2003). ITS secondary structure derived from comparative ana- lysis: implications for sequence alignment and phylogeny of the Asteraceae.Mol Phylogenet Evol 29, 216-234. |
| 16 | Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Regev A (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome.Nat Biotechnol 29, 644-652. |
| 17 | Jansen RK, Cai Z, Raubeson LA, Daniell H, Leebens- Mack J, Müller KF, Boore JL (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns.Proc Natl Acad Sci USA 104, 19369-19374. |
| 18 | Jansen RK, Holsinger KE, Michaels HJ, Palmer JD (1990). Phylogenetic analysis of chloroplast DNA restric- tion site data at higher taxonomic levels: an example from the Asteraceae.Evolution 44, 2089-2105. |
| 19 | Jansen RK, Michaels HJ, Palmer JD (1991). Phylogeny and character evolution in the Asteraceae based on chloroplast DNA restriction site mapping.Syst Bot 16, 98-115. |
| 20 | Karis PO (1993). Morphological phylogenetics of the As- teraceae-Asteroideae, with notes on character evolution.Plant Syst Evol 186, 69-93. |
| 21 | Karis PO (1994). Phylogeny of Asteraceae: Asteroideae revisited. In: Hind DJN, Beentje HJ, eds. Compositae: Systematics. Kew: Royal Botanic Gardens Publishers. pp. 41-47. |
| 22 | Karis PO, Ryding O (1994). Tribe heliantheae. In: Bremer KE, ed. Asteraceae, Cladistics & Classification. Portland: Timber Press. pp. 559-624. |
| 23 | Kim KJ, Jansen RK (1995). ndhF sequence evolution and the major clades in the sunflower family.Proc Natl Acad Sci USA 92, 10379-10383. |
| 24 | Kim KJ, Jasen RK, Wallace RS, Michaels HJ, Palmer JD (1992). Phylogenetic implications of rbcL sequence varia- tion in the Asteraceae.Ann Mo Bot Gard 79, 428-445. |
| 25 | Lin ZG, Kong HZ, Nei M, Ma H (2006). Origins and evolu- tion of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer.Proc Natl Acad Sci USA 103, 10328-10333. |
| 26 | Liu JQ, Gao TG, Chen ZD, Lu AM (2002). Molecular phylogeny and biogeography of the Qinghai-Tibet Plateau endemic Nannoglottis (Asteraceae).Mol Phylogenet Evol 23, 307-325. |
| 27 | Liu L, Wu SY, Yu LL (2015). Coalescent methods for estimating species trees from phylogenomic data.J Syst Evol 53, 380-390. |
| 28 | Lockhart PJ, Penny D (2005). The place of Amborella within the radiation of angiosperms.Trends Plant Sci 10, 201-202. |
| 29 | Mandel JR, Dikow RB, Funk VA (2015). Using phylogenomics to resolve mega-families: an example from Compositae.J Syst Evol 53, 391-402. |
| 30 | Mandel JR, Dikow RB, Funk VA, Masalia RR, Staton SE, Kozik A, Burke JM (2014). A target enrichment method for gathering phylogenetic information from hundreds of loci: an example from the Compositae.Appl Plant Sci 2, 130085. |
| 31 | Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T (2014). ASTRAL: genome-scale coales- cent-based species tree estimation.Bioinformatics 30, 541-548. |
| 32 | Moore MJ, Bell CD, Soltis PS, Soltis DE (2007). Using plastid genome-scale data to resolve enigmatic relation- ships among basal angiosperms.Proc Natl Acad Sci USA 104, 19363-19368. |
| 33 | Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots.Proc Natl Acad Sci USA 107, 4623-4628. |
| 34 | Nesom GL, Robinson H (2007). Astereae. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 284-342. |
| 35 | Ness RW, Graham SW, Barrett SC (2011). Reconciling gene and genome duplication events: using multiple nuclear gene families to infer the phylogeny of the aquatic plant family Pontederiaceae.Mol Biol Evol 28, 3009-3018. |
| 36 | Ng M, Yanofsky MF (2001). Function and evolution of the plant MADS-box gene family.Nat Rev Genet 2, 186-195. |
| 37 | Nordenstam B, Källersjö M (2009). Calenduleae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 527-538. |
| 38 | Nordenstam B, Pelser PB, Kadereit JW, Watson LE (2009). Senecioneae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 503-526. |
| 39 | Oberprieler C, Himmelreich S, Källersjö M, Vallès J, Watson LE, Vogt R (2009). Anthemideae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxonomy. pp. 631-666. |
| 40 | Oberprieler C, Vogt R, Watson LE (2007). Anthemideae. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 243-374. |
| 41 | Panero JL (2005). New combinations and infrafamilial taxa in the Asteraceae. Phytologia 87, 1-14. |
| 42 | Panero JL (2007a). Key to the tribes of the Heliantheae alliance. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 391-395. |
| 43 | Panero JL (2007b). Tribe Coreopsideae. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 406-417. |
| 44 | Panero JL (2007c). Tribe Millerieae. In: Kadereit JW, Jeffrey C, eds. The Families and Genera of Vascular Plants, Vol. 8, Flowering Plants. Eudicots. Asterales. Berlin: Springer. pp. 477-492. |
| 45 | Panero JL, Freire SE, Espinar LA, Crozier BS, Barboza GE, Cantero JJ (2014). Resolution of deep nodes yields an improved backbone phylogeny and a new basal lineage to study early evolution of Asteraceae.Mol Phylogenet Evol 80, 43-53. |
| 46 | Panero JL, Funk VA (2002). Toward a phylogenetic subfamilial classification for the Compositae (Asteraceae).P Biol Soc Wash 115, 909-922. |
| 47 | Panero JL, Funk VA (2008). The value of sampling ano- malous taxa in phylogenetic studies: major clades of the Asteraceae revealed.Mol Phylogenet Evol 47, 757-782. |
| 48 | Pelser PB, Kennedy AH, Tepe EJ, Shidler JB, Nordenstam B, Kadereit JW, Watson LE (2010). Patterns and causes of incongruence between plastid and nuclear Senecioneae (Asteraceae) phylogenies.Am J Bot 97, 856-873. |
| 49 | Pelser PB, Watson L (2009). Introduction to Asteroideae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, eds. Systematics, Evolution, and Biogeography of Compositae. Vienna: International Association for Plant Taxanomy. pp. 495-502. |
| 50 | Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Quackenbush J (2003). TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets.Bioinformatics 19, 651-652. |
| 51 | Phillips MJ, Delsuc F, Penny D (2004). Genome-scale phylogeny and the detection of systematic biases.Mol Biol Evol 21, 1455-1458. |
| 52 | Rieseberg LH, Soltis DE (1991). Phylogenetic consequences of cytoplasmic gene flow in plants.Evol Trends Plants 5, 65-84. |
| 53 | Robinson H (1981). A revision of the tribal and subtribal limits of the Heliantheae (Asteraceae).Smithsonian Contrib Bot 51, 1-102. |
| 54 | Robinson H (2004). New supertribes, Helianthodae and Senecionodae, for the subfamily Asteroideae (Asterac- eae).Phytologia 86, 116-120. |
| 55 | Robinson H (2005). Validation of the supertribe Asterodae.Phytologia 87, 73-74. |
| 56 | Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models.Bioinformatics 19, 1572-1574. |
| 57 | Sang T, Zhong Y (2000). Testing hybridization hypotheses based on incongruent gene trees.Syst Biol 49, 422-434. |
| 58 | Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Soltis PS (2009). Polyploidy and angiosperm diversification.Am J Bot 96, 336-348. |
| 59 | Soltis DE, Albert VA, Savolainen V, Hilu K, Qiu YL, Chase MW, Soltis PS (2004). Genome-scale data, angiosperm relationships, and ‘ending incongruence’: a cautionary tale in phylogenetics.Trends Plant Sci 9, 477-483. |
| 60 | Soltis DE, Kuzoff RK (1995). Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxi- fragaceae).Evolution 49, 727-742. |
| 61 | Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models.Bioinformatics 22, 2688-2690. |
| 62 | Stefanović S, Rice DW, Palmer JD (2004). Long branch attraction, taxon sampling, and the earliest angiosperms: amborella or monocots?BMC Evol Biol 4, 35. |
| 63 | Stuessy TF (1977). Heliantheae-systematic review. In: Hey- wood VH, Harborne JB, Turner BL, eds. The Biology and Hemistry of the Compositae. London: Academic Press. pp. 621-671. |
| 64 | Suyama M, Torrents D, Bork P (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments.Nucleic Acids Res 34, W609-W612. |
| 65 | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: molecular evolutionary genetics analysis version 6.0.Mol Biol Evol 30, 2725-2729. |
| 66 | Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Leebens-Mack J (2014). Phylotranscrip- tomic analysis of the origin and early diversification of land plants.Proc Natl Acad Sci USA 111, E4859-E4868. |
| 67 | Xu GX, Ma H, Nei M, Kong HZ (2009). Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification.Proc Natl Acad Sci USA 106, 835-840. |
| 68 | Yang Y, Moore MJ, Brockington SF, Soltis DE, Wong GKS, Carpenter EJ, Smith SA (2015). Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing.Mol Biol Evol (in press) |
| 69 | Zeng LP, Zhang Q, Sun RR, Kong HZ, Zhang N, Ma H (2014). Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times.Nat Commun 5, 4956. |
| 70 | Zimmer EA, Wen J (2012). Using nuclear gene data for plant phylogenetics: progress and prospects.Mol Phylogenet Evol 65, 774-785. |
| 71 | Zimmer EA, Wen J (2015). Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches.J Syst Evol 53, 371-379. |
| [1] | Hong Deyuan. A brief discussion on methodology in taxonomy [J]. Biodiv Sci, 2025, 33(2): 24541-. |
| [2] | Yajun Sun. What do higher or lower organisms mean—Clarify the meaning and validity of the biological ladder implied by On the Origin of Species [J]. Biodiv Sci, 2025, 33(1): 24394-. |
| [3] | Hua He, Dunyan Tan, Xiaochen Yang. Cryptic dioecy in angiosperms: Diversity, phylogeny and evolutionary significance [J]. Biodiv Sci, 2024, 32(6): 24149-. |
| [4] | Yanyu Ai, Haixia Hu, Ting Shen, Yuxuan Mo, Jinhua Qi, Liang Song. Vascular epiphyte diversity and the correlation analysis with host tree characteristics: A case in a mid-mountain moist evergreen broad-leaved forest, Ailao Mountains [J]. Biodiv Sci, 2024, 32(5): 24072-. |
| [5] | Yanwen Lv, Ziyun Wang, Yu Xiao, Zihan He, Chao Wu, Xinsheng Hu. Advances in lineage sorting theories and their detection methods [J]. Biodiv Sci, 2024, 32(4): 23400-. |
| [6] | Zhi Yang, Yong Yang. Research Advances on Nuclear Genomes of Economically Important Trees of Lauraceae [J]. Chinese Bulletin of Botany, 2024, 59(2): 302-318. |
| [7] | Chen-Kun Jiang, Wen-Bin Yu, Guang-Yuan Rao, Huaicheng Li, Julien B. Bachelier, Hartmut H. Hilger, Theodor C. H. Cole. Plant Phylogeny Posters—An educational project on plant diversity from an evolutionary perspective [J]. Biodiv Sci, 2024, 32(11): 24210-. |
| [8] | Zhenzhou Chu, Gulbar Yisilam, Zezhong Qu, Xinmin Tian. Comparative Analyses on the Chloroplast Genome of Three Sympatric Atraphaxis Species [J]. Chinese Bulletin of Botany, 2023, 58(3): 417-432. |
| [9] | Huiyin Song, Zhengyu Hu, Guoxiang Liu. Assessing advances in taxonomic research on Chlorellaceae (Chlorophyta) [J]. Biodiv Sci, 2023, 31(2): 22083-. |
| [10] | Jinbo Bao, Zhijie Ding, Haoyu Miao, Xueli Li, Shuxian Ren, Ruoyan Jiao, Hao Li, Qianqian Deng, Yingzi Li, Xinmin Tian. Analysis of Chloroplast Genomes of Aleurites moluccana [J]. Chinese Bulletin of Botany, 2023, 58(2): 248-260. |
| [11] | Zhizhong Li, Shuai Peng, Qingfeng Wang, Wei Li, Shichu Liang, Jinming Chen. Cryptic diversity of the genus Ottelia in China [J]. Biodiv Sci, 2023, 31(2): 22394-. |
| [12] | Ting Wang, Jiangping Shu, Yufeng Gu, Yanqing Li, Tuo Yang, Zhoufeng Xu, Jianying Xiang, Xianchun Zhang, Yuehong Yan. Insight into the studies on diversity of lycophytes and ferns in China [J]. Biodiv Sci, 2022, 30(7): 22381-. |
| [13] | Jianming Wang, Mengjun Qu, Yin Wang, Yiming Feng, Bo Wu, Qi Lu, Nianpeng He, Jingwen Li. The drivers of plant taxonomic, functional, and phylogenetic β-diversity in the gobi desert of northern Qinghai-Tibet Plateau [J]. Biodiv Sci, 2022, 30(6): 21503-. |
| [14] | XIANG Wei, HUANG Dong-Liu, ZHU Shi-Dan. Absorptive root anatomical traits of 26 tropical and subtropical fern species [J]. Chin J Plant Ecol, 2022, 46(5): 593-601. |
| [15] | WANG Chun-Cheng, ZHANG Yun-Ling, MA Song-Mei, HUANG Gang, ZHANG Dan, YAN Han. Phylogeny and species differentiation of four wild almond species of subgen. Amygdalus in China [J]. Chin J Plant Ecol, 2021, 45(9): 987-995. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||