

Chinese Bulletin of Botany ›› 2022, Vol. 57 ›› Issue (5): 611-622.DOI: 10.11983/CBB22046 cstr: 32102.14.CBB22046
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Lin Haijiao, Qu Jiaqi, Liu Yinan, Yuan Zening*( )
)
Received:2022-03-10
Accepted:2022-05-30
Online:2022-09-01
Published:2022-09-09
Contact:
Yuan Zening
About author:*E-mail: xiaoyuan168ok@163.comLin Haijiao, Qu Jiaqi, Liu Yinan, Yuan Zening. Physiological and Molecular Response Mechanisms Under Low-temperature Stress in Lavandula angustifolia Leaves[J]. Chinese Bulletin of Botany, 2022, 57(5): 611-622.
| Gene ID | Gene name | Primer sequence (5′-3′) | |
|---|---|---|---|
| DN12834_c2_g2 | LaLEA1 | F: CCAAGGCAATCTCTGCTCTCA | R: CTTTGGATCCGGAGCCTTTCT |
| DN11187_c0_g4 | LaLEA2 | F: TCTACGTCTTCATCTTCGCCG | R: GCGTTGTAGTTTGCCAGAGTG |
| DN10094_c0_g1 | LaERD10 | F: ATATGACGACGCACCGACTG | R: TCCGGTGCGTAATCCGAATC |
| DN14123_c1_g1 | LaFAB2 | F: CGACTTTTGTCTCCCACGGA | R: TGTCCCATCTGGGTCGATCT |
| DN16955_c0_g1 | LaPOD2 | F: CATGGACTATGTCACGCCGA | R: GCTCGAAGAAAGCAATGGCA |
| DN18853_c0_g2 | LaBAM1 | F: CCCACGGCGAGAGAATTGTA | R: TTTGAGCGATGGGGACGTAG |
| GADPH | F: TAGGAGGTGGCAGGACATCA | R: CCCTTTACCCGTCACGTTGT | |
Table 1 Primers used for qRT-PCR
| Gene ID | Gene name | Primer sequence (5′-3′) | |
|---|---|---|---|
| DN12834_c2_g2 | LaLEA1 | F: CCAAGGCAATCTCTGCTCTCA | R: CTTTGGATCCGGAGCCTTTCT |
| DN11187_c0_g4 | LaLEA2 | F: TCTACGTCTTCATCTTCGCCG | R: GCGTTGTAGTTTGCCAGAGTG |
| DN10094_c0_g1 | LaERD10 | F: ATATGACGACGCACCGACTG | R: TCCGGTGCGTAATCCGAATC |
| DN14123_c1_g1 | LaFAB2 | F: CGACTTTTGTCTCCCACGGA | R: TGTCCCATCTGGGTCGATCT |
| DN16955_c0_g1 | LaPOD2 | F: CATGGACTATGTCACGCCGA | R: GCTCGAAGAAAGCAATGGCA |
| DN18853_c0_g2 | LaBAM1 | F: CCCACGGCGAGAGAATTGTA | R: TTTGAGCGATGGGGACGTAG |
| GADPH | F: TAGGAGGTGGCAGGACATCA | R: CCCTTTACCCGTCACGTTGT | |
| Levels of chilling injury | Standard of judgment |
|---|---|
| 1 | Leaf present green and there are no symptoms of chilling injury |
| 2 | Only the edge of the leaf is yellow |
| 3 | Leaf withered and yellow part of < 50%, the green part is more |
| 4 | 50%-100% of the leaf is dry and yellow, with less green |
| 5 | The whole plant is wilting |
| 6 | The aboveground parts dried up and died |
Table 2 Symptoms grading of plant chilling injury
| Levels of chilling injury | Standard of judgment |
|---|---|
| 1 | Leaf present green and there are no symptoms of chilling injury |
| 2 | Only the edge of the leaf is yellow |
| 3 | Leaf withered and yellow part of < 50%, the green part is more |
| 4 | 50%-100% of the leaf is dry and yellow, with less green |
| 5 | The whole plant is wilting |
| 6 | The aboveground parts dried up and died |
| Database | Number of annotated unigene | Percentage of annotated unigene (%) |
|---|---|---|
| NT | 32797 | 42.73 |
| NR | 48554 | 63.26 |
| Uniprot (BLASTX) | 37801 | 49.25 |
| Uniprot (BLASTP) | 25631 | 33.39 |
| PFAM | 22340 | 29.11 |
| eggNOG | 28488 | 37.12 |
| GO | 24430 | 31.83 |
| KEGG | 15496 | 20.19 |
| KOG | 28787 | 37.51 |
Table 3 The statistical result of unigene functional annotation
| Database | Number of annotated unigene | Percentage of annotated unigene (%) |
|---|---|---|
| NT | 32797 | 42.73 |
| NR | 48554 | 63.26 |
| Uniprot (BLASTX) | 37801 | 49.25 |
| Uniprot (BLASTP) | 25631 | 33.39 |
| PFAM | 22340 | 29.11 |
| eggNOG | 28488 | 37.12 |
| GO | 24430 | 31.83 |
| KEGG | 15496 | 20.19 |
| KOG | 28787 | 37.51 |
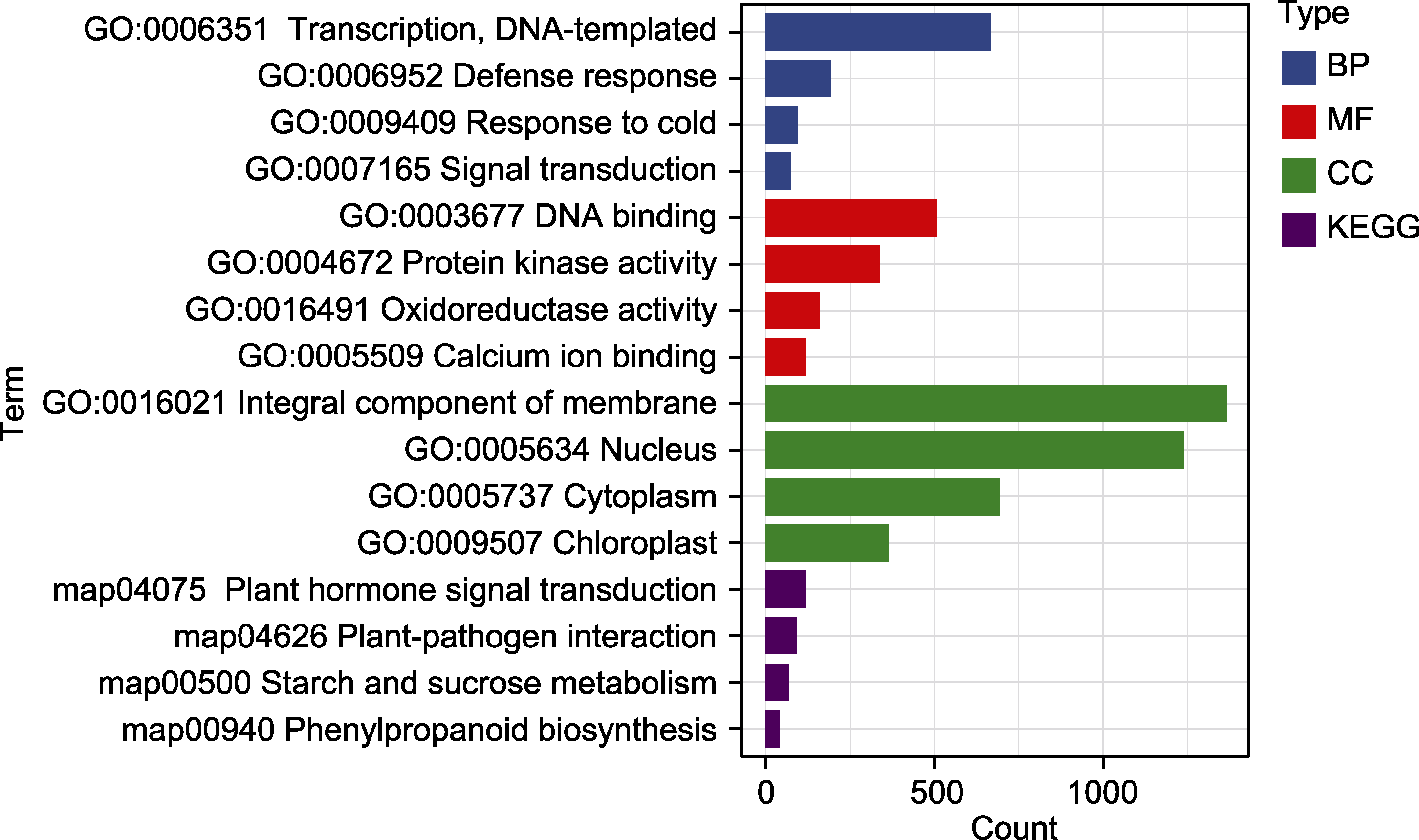
Figure 2 GO and KEGG enrichment of DEGs in Lavandula angustifolia leaves under low-temperature stress (0°C vs 20°C) BP: Biological process; MF: Molecular function; CC: Cellular component
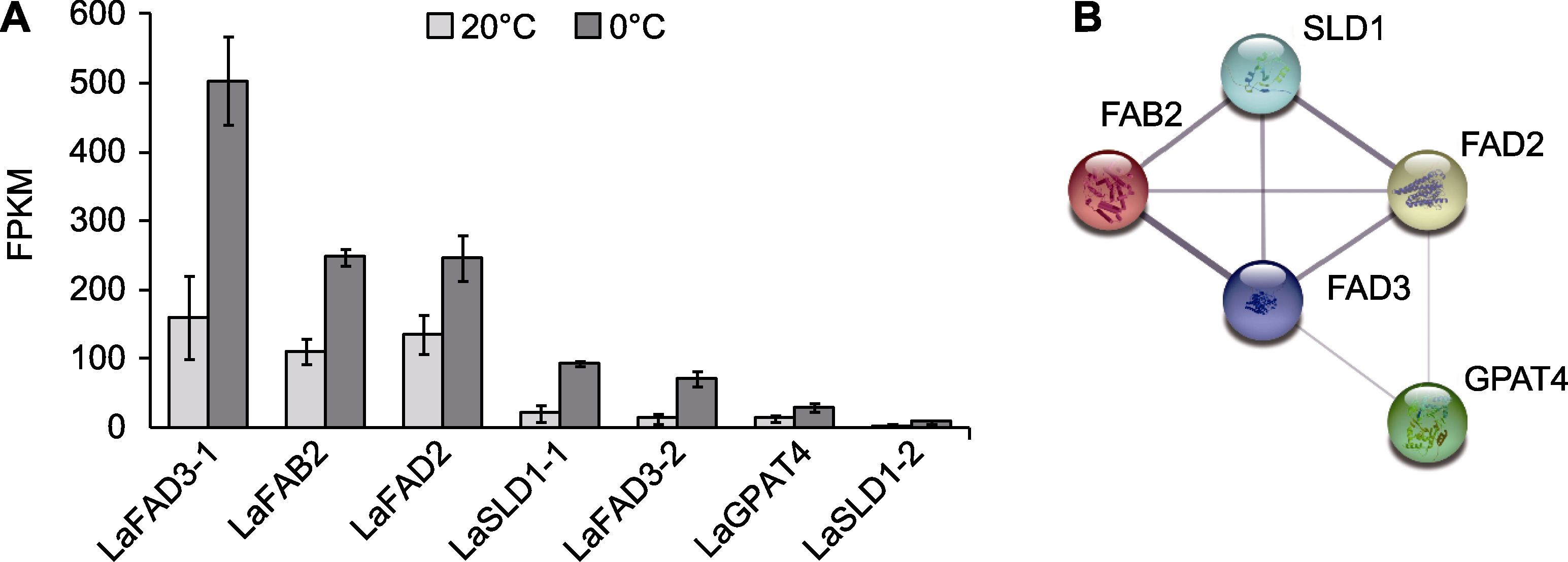
Figure 3 Changes in expression of cold tolerance genes related to membrane stability of Lavandula angustifolia and the interaction network of these encoding proteins (A) Gene expression changes (0°C vs 20°C); (B) Protein interaction network. FPKM: Fragments per kilobase per million mapped fragments
| Gene ID | Gene name | Number of amino acids (aa) | Molecular weight (kDa) | Theoretical pI | Grand average of hydropathicity (GRAVY) | Aliphatic index | Instability index | Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| DN12834_c2_g2 | LaLEA1 | 100 | 10.90 | 10.32 | -0.484 | 73.40 | 52.96 | - |
| DN11187_c0_g4 | LaLEA2 | 128 | 14.16 | 10.09 | -0.088 | 74.77 | 22.26 | Chloroplasts |
| DN13720_c1_g4 | LaLEA3 | 264 | 28.68 | 5.12 | -0.933 | 59.73 | 14.27 | Chloroplasts |
| DN9448_c0_g7 | LaLEA4 | 193 | 21.28 | 10.13 | 0.123 | 92.49 | 43.01 | Chloroplasts |
| DN10042_c1_g5 | LaLEA5 | 176 | 19.78 | 10.52 | -0.463 | 83.01 | 34.04 | Chloroplasts |
| DN20068_c2_g4 | LaLEA6 | 122 | 13.28 | 10.07 | -0.204 | 71.15 | 35.17 | - |
| DN13618_c0_g1 | LaLEA7 | 120 | 13.63 | 9.68 | -0.299 | 61.08 | 16.35 | Nucleus |
| DN7134_c0_g1 | LaLEA8 | 208 | 22.93 | 9.97 | -0.062 | 92.74 | 28.05 | - |
| DN16104_c0_g1 | LaLEA9 | 219 | 24.60 | 9.97 | -0.357 | 72.19 | 41.89 | - |
| DN9448_c0_g2 | LaLEA10 | 132 | 14.84 | 10.06 | -0.475 | 67.42 | 24.52 | - |
| DN19299_c2_g2 | LaLEA11 | 320 | 35.11 | 9.63 | -0.674 | 55.44 | 89.14 | - |
| DN11058_c1_g6 | LaLEA12 | 124 | 13.00 | 8.83 | -1.085 | 41.94 | 29.73 | Mitochondrial |
| DN17935_c0_g1 | LaLEA13 | 228 | 25.24 | 10.37 | -0.224 | 89.74 | 42.60 | - |
| DN5364_c0_g1 | LaLEA14 | 229 | 25.76 | 9.85 | -0.035 | 90.66 | 40.29 | - |
| DN13618_c0_g3 | LaLEA15 | 134 | 14.75 | 10.01 | -0.440 | 64.03 | 24.39 | - |
| DN24875_c0_g1 | LaLEA16 | 121 | 12.51 | 8.78 | -1.135 | 31.98 | 32.24 | Nucleus |
| DN9962_c0_g1 | LaLEA17 | 225 | 24.75 | 9.69 | -0.036 | 98.67 | 35.53 | Cytoplasm |
| DN10042_c1_g1 | LaLEA18 | 203 | 22.61 | 10.07 | -0.226 | 95.02 | 30.10 | - |
| DN11740_c4_g1 | LaLEA19 | 133 | 13.80 | 4.64 | -0.514 | 72.56 | 19.99 | Chloroplasts |
| DN11740_c4_g1 | LaLEA20 | 356 | 37.92 | 5.88 | -1.015 | 52.16 | 17.85 | Chloroplasts |
| DN4252_c0_g1 | LaLEA21 | 259 | 26.30 | 4.89 | -0.167 | 82.43 | 30.36 | Cytoplasm |
| DN6726_c0_g1 | LaLEA22 | 204 | 22.22 | 10.42 | -0.135 | 88.97 | 40.07 | - |
| DN10094_c0_g1 | LaERD10 | 155 | 17.51 | 5.79 | -1.632 | 45.94 | 48.98 | - |
Table 4 Physical and chemical information of LaLEAs of Lavandula angustifolia
| Gene ID | Gene name | Number of amino acids (aa) | Molecular weight (kDa) | Theoretical pI | Grand average of hydropathicity (GRAVY) | Aliphatic index | Instability index | Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| DN12834_c2_g2 | LaLEA1 | 100 | 10.90 | 10.32 | -0.484 | 73.40 | 52.96 | - |
| DN11187_c0_g4 | LaLEA2 | 128 | 14.16 | 10.09 | -0.088 | 74.77 | 22.26 | Chloroplasts |
| DN13720_c1_g4 | LaLEA3 | 264 | 28.68 | 5.12 | -0.933 | 59.73 | 14.27 | Chloroplasts |
| DN9448_c0_g7 | LaLEA4 | 193 | 21.28 | 10.13 | 0.123 | 92.49 | 43.01 | Chloroplasts |
| DN10042_c1_g5 | LaLEA5 | 176 | 19.78 | 10.52 | -0.463 | 83.01 | 34.04 | Chloroplasts |
| DN20068_c2_g4 | LaLEA6 | 122 | 13.28 | 10.07 | -0.204 | 71.15 | 35.17 | - |
| DN13618_c0_g1 | LaLEA7 | 120 | 13.63 | 9.68 | -0.299 | 61.08 | 16.35 | Nucleus |
| DN7134_c0_g1 | LaLEA8 | 208 | 22.93 | 9.97 | -0.062 | 92.74 | 28.05 | - |
| DN16104_c0_g1 | LaLEA9 | 219 | 24.60 | 9.97 | -0.357 | 72.19 | 41.89 | - |
| DN9448_c0_g2 | LaLEA10 | 132 | 14.84 | 10.06 | -0.475 | 67.42 | 24.52 | - |
| DN19299_c2_g2 | LaLEA11 | 320 | 35.11 | 9.63 | -0.674 | 55.44 | 89.14 | - |
| DN11058_c1_g6 | LaLEA12 | 124 | 13.00 | 8.83 | -1.085 | 41.94 | 29.73 | Mitochondrial |
| DN17935_c0_g1 | LaLEA13 | 228 | 25.24 | 10.37 | -0.224 | 89.74 | 42.60 | - |
| DN5364_c0_g1 | LaLEA14 | 229 | 25.76 | 9.85 | -0.035 | 90.66 | 40.29 | - |
| DN13618_c0_g3 | LaLEA15 | 134 | 14.75 | 10.01 | -0.440 | 64.03 | 24.39 | - |
| DN24875_c0_g1 | LaLEA16 | 121 | 12.51 | 8.78 | -1.135 | 31.98 | 32.24 | Nucleus |
| DN9962_c0_g1 | LaLEA17 | 225 | 24.75 | 9.69 | -0.036 | 98.67 | 35.53 | Cytoplasm |
| DN10042_c1_g1 | LaLEA18 | 203 | 22.61 | 10.07 | -0.226 | 95.02 | 30.10 | - |
| DN11740_c4_g1 | LaLEA19 | 133 | 13.80 | 4.64 | -0.514 | 72.56 | 19.99 | Chloroplasts |
| DN11740_c4_g1 | LaLEA20 | 356 | 37.92 | 5.88 | -1.015 | 52.16 | 17.85 | Chloroplasts |
| DN4252_c0_g1 | LaLEA21 | 259 | 26.30 | 4.89 | -0.167 | 82.43 | 30.36 | Cytoplasm |
| DN6726_c0_g1 | LaLEA22 | 204 | 22.22 | 10.42 | -0.135 | 88.97 | 40.07 | - |
| DN10094_c0_g1 | LaERD10 | 155 | 17.51 | 5.79 | -1.632 | 45.94 | 48.98 | - |
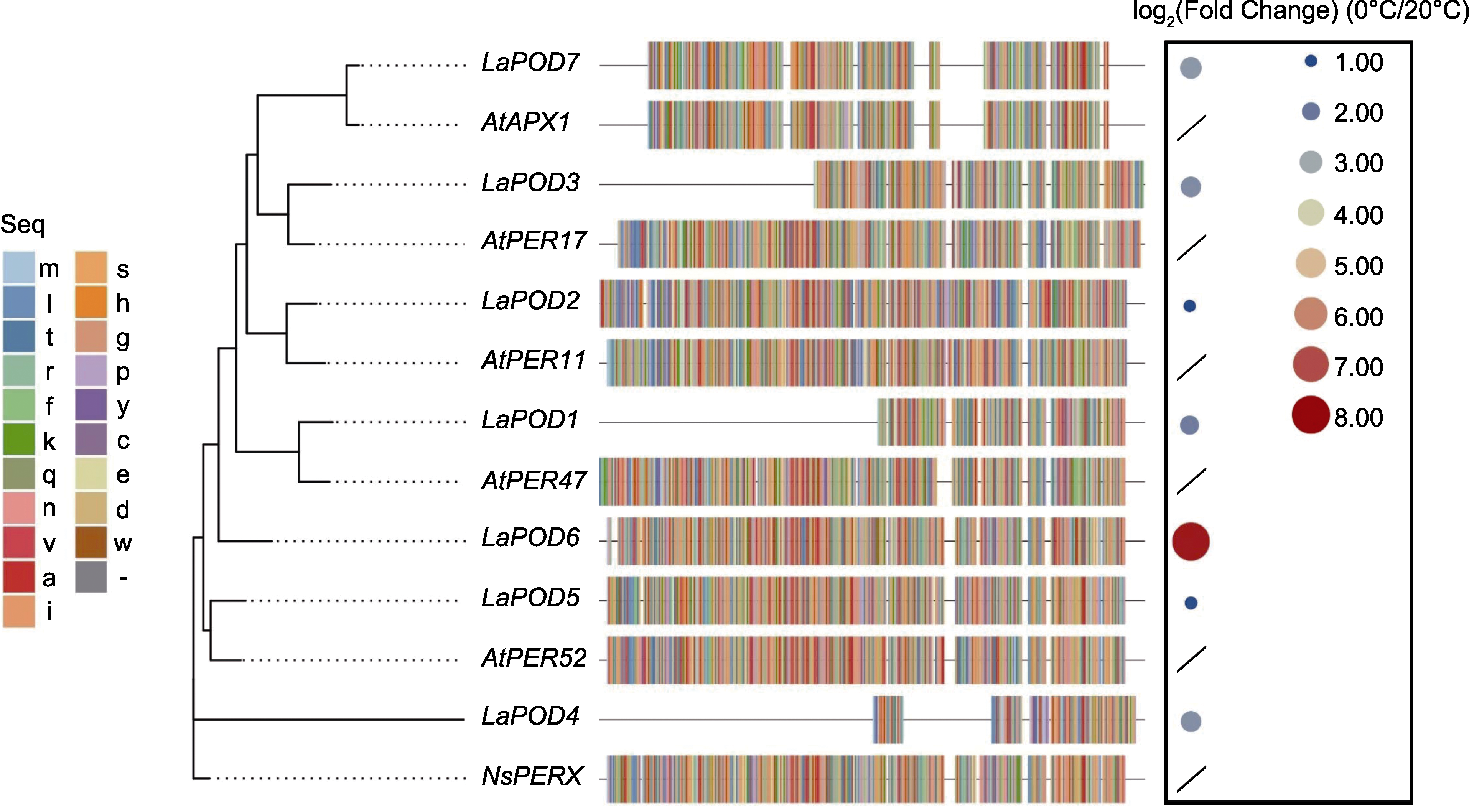
Figure 6 The evolutionary tree of Lavandula angustifolia and it’s relative species, and LaPODs expression changes under low-temperature stress (0°C vs 20°C)
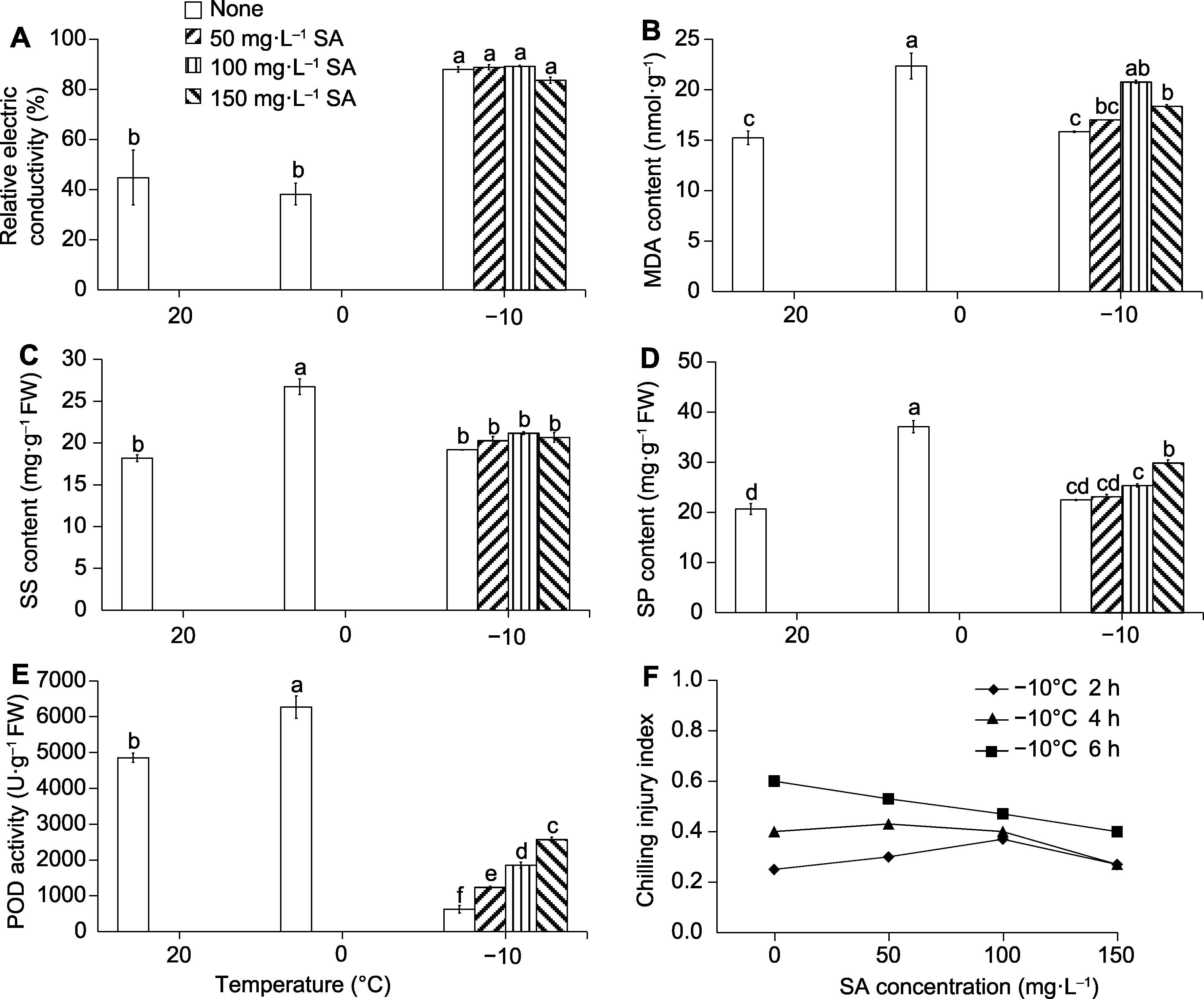
Figure 8 Physiological changes of Lavandula angustifolia leaves under chilling and freezing conditions (A-E) and alleviation of freezing injury by salicylic acid (SA) (F) MDA: Malondialdehyde; SS: Soluble sugars; SP: Soluble proteins; POD: Peroxidase. Different lowercase letters indicate significant differences among different treatments (P<0.05).
| [1] | 陈建权, 程晨, 张梦恬, 张向前, 张尧, 王爱英, 祝建波 (2018). 天山雪莲SiSAD基因与拟南芥AtFAB2基因转化烟草的抗寒性分析. 植物学报 53, 603-611. |
| [2] | 代宇佳, 罗晓峰, 周文冠, 陈锋, 帅海威, 杨文钰, 舒凯 (2019). 生物和非生物逆境胁迫下的植物系统信号. 植物学报 54, 255-264. |
| [3] | 段志坤, 秦晓惠, 朱晓红, 宋纯鹏 (2018). 解析植物冷信号转导途径: 植物如何感知低温. 植物学报 53, 149-153. |
| [4] | 何子华, 杨成行, 王沛, 包爱科, 马清 (2021). 高寒地区6种禾本科牧草对低温胁迫的生理响应及耐寒性评价. 草业科学 38, 2019-2028. |
| [5] | 李瑞雪, 金晓玲, 胡希军, 汪结明, 罗峰, 张方静 (2019). 低温胁迫下6种木兰科植物的生理响应及抗寒相关基因差异表达. 生态学报 39, 2883-2898. |
| [6] | 王笑, 蔡剑, 周琴, 戴廷波, 姜东 (2021). 非生物逆境锻炼提高作物耐逆性的生理机制研究进展. 中国农业科学 54, 2287-2301. |
| [7] | 张超 (2018). 大豆FAD3基因的克隆及其功能验证. 硕士论文. 长春: 吉林大学. pp. 41-42. |
| [8] | Al-Ansari MM, Andeejani AMI, Alnahmi E, AlMalki RH, Masood A, Vijayaraghavan P, Rahman AA, Choi KC (2021). Insecticidal, antimicrobial and antioxidant activities of essential oil from Lavandula latifolia L. and its deterrent effects on Euphoria leucographa. Ind Crops Prod 170, 113740. |
| [9] | Barrero-Gil J, Salinas J (2018). Gene regulatory networks mediating cold acclimation: the CBF pathway. In: Iwaya- Inoue M, Sakurai M, Uemura M, eds. Survival Strategies in Extreme Cold and Desiccation. Singapore: Springer. pp. 3-22. |
| [10] | Chinnusamy V, Zhu JK (2009). Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12, 133-139. |
| [11] | Chrysargyris A, Laoutari S, Litskas VD, Stavrinides MC, Tzortzakis N (2016). Effects of water stress on lavender and sage biomass production, essential oil composition and biocidal properties against Tetranychus urticae (Koch). Sci Hortic 213, 96-103. |
| [12] | Chrysargyris A, Michailidi E, Tzortzakis N (2018). Physiological and biochemical responses of Lavandula angustifolia to salinity under mineral foliar application. Front Plant Sci 9, 489. |
| [13] | Ding YL, Shi YT, Yang SH (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol 222, 1690-1704. |
| [14] | Ding YL, Shi YT, Yang SH (2020). Molecular regulation of plant responses to environmental temperatures. Mol Plant 13, 544-564. |
| [15] | Du B, Rennenberg H (2018). Physiological responses of lavender (Lavandula angustifolia Mill.) to water deficit and recovery. S Afr J Bot 119, 212-218. |
| [16] | Duan XJ, Zhu ZL, Yang Y, Duan J, Jia ZK, Chen FJ, Sang ZY, Ma LY (2022). Salicylic acid regulates sugar metabolism that confers freezing tolerance in Magnolia wufengensis during natural cold acclimation. J Plant Growth Regul 41, 227-235. |
| [17] | Golizadeh F, Kumleh HH (2019). Physiological responses and expression changes of fatty acid metabolism-related genes in wheat (Triticum aestivum) under cold stress. Plant Mol Biol Rep 37, 224-236. |
| [18] | Guo XY, Liu DF, Chong K (2018). Cold signaling in plants: insights into mechanisms and regulation. J Integr Plant Biol 60, 745-756. |
| [19] | Guy CL, Niemi KJ, Brambl R (1985). Altered gene expression during cold acclimation of spinach. Proc Natl Acad Sci USA 82, 3673-3677. |
| [20] | Ignatenko A, Talanova V, Repkina N, Titov A (2019). Exogenous salicylic acid treatment induces cold tolerance in wheat through promotion of antioxidant enzyme activity and proline accumulation. Acta Physiol Plant 41, 80. |
| [21] | John R, Anjum NA, Sopory SK, Akram NA, Ashraf M (2016). Some key physiological and molecular processes of cold acclimation. Biol Plant 60, 603-618. |
| [22] | Kargiotidou A, Deli D, Galanopoulou D, Tsaftaris A, Farmaki T (2008). Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J Exp Bot 59, 2043-2056. |
| [23] | Li JR, Wang YM, Dong YM, Zhang WY, Wang D, Bai HT, Li K, Li H, Shi L (2021). The chromosome-based lavender genome provides new insights into Lamiaceae evolution and terpenoid biosynthesis. Hortic Res 8, 90. |
| [24] | Li XT, Liu P, Yang PP, Fan CZ, Sun XM (2018). Characterization of the glycerol-3-phosphate acyltransferase gene and its real-time expression under cold stress in Paeonia lactiflora Pall. PLoS One 13, e0202168. |
| [25] | Liu J, Li JM, Fu CX (2021). Comparative physiology and transcriptome analysis reveals the regulatory mechanism of genome duplication enhancing cold resistance in Fragaria nilgerrensis. Environ Exp Bot 188, 104509. |
| [26] | Liu JY, Shi YT, Yang SH (2018). Insights into the regulation of C-repeat binding factors in plant cold signaling. J Integr Plant Biol 60, 780-795. |
| [27] | Paraskevopoulou AT, Tsarouchas P, Londra PA, Kamoutsis AP (2020). The effect of irrigation treatment on the growth of lavender species in an extensive green roof system. Water 12, 863. |
| [28] | Pedrosa AM, de Paula Santos Martins C, Gonçalves LP, Costa MGC (2015). Late embryogenesis abundant (LEA) constitutes a large and diverse family of proteins involved in development and abiotic stress responses in sweet orange (Citrus sinensis L. Osb.). PLoS One 10, e0145785. |
| [29] | Peng YJ, Yang JF, Li X, Zhang YL (2021). Salicylic acid: biosynthesis and signaling. Annu Rev Plant Biol 72, 761-791. |
| [30] | Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996). Systemic acquired resistance. Plant Cell 8, 1809-1819. |
| [31] | Shi HF, He XY, Zhao YJ, Lu SY, Guo ZF (2020). Constitutive expression of a group 3 LEA protein from Medicago falcata (MfLEA3) increases cold and drought tolerance in transgenic tobacco. Plant Cell Rep 39, 851-860. |
| [32] | Szekely-Varga Z, González-Orenga S, Cantor M, Jucan D, Boscaiu M, Vicente O (2020). Effects of drought and salinity on two commercial varieties of Lavandula angustifolia Mill. Plants 9, 637. |
| [33] | Wang WL, Wang X, Huang M, Cai J, Zhou Q, Dai TB, Cao WX, Jiang D (2018). Hydrogen peroxide and abscisic acid mediate salicylic acid-induced freezing tolerance in wheat. Front Plant Sci 9, 1137. |
| [34] | Wang X, Yu C, Liu Y, Yang L, Li Y, Yao W, Cai YC, Yan X, Li SB, Cai YH, Li SQ, Peng XJ (2019). GmFAD3A, A ω-3 fatty acid desaturase gene, enhances cold tolerance and seed germination rate under low temperature in rice. Int J Mol Sci 20, 3796. |
| [35] | Wang Y, Li Y, Wang JH, Xiang Z, Xi PY, Zhao DG (2021). Physiological changes and differential gene expression of tea plants (Camellia sinensis (L.) Kuntze var. niaowangensis Q. H. Chen) under cold stress. DNA Cell Biol 40, 906-920. |
| [36] | Wu QY, He TJ, Liu H, Luo XB, Yin W, Chen EF, Li F (2019). Cell ultrastructure and physiological changes of potato during cold acclimation. Can J Plant Sci 99, 873-884. |
| [37] | Xu ML, Tong Q, Wang Y, Wang ZM, Xu GZ, Elias GK, Li SH, Liang ZC (2020). Transcriptomic analysis of the grapevine LEA gene family in response to osmotic and cold stress reveals a key role for VamDHN3. Plant Cell Physiol 61, 775-786. |
| [38] | Xu XX, Yan BW, Zhao Y, Wang F, Zhao XC, He L, Xu JY, Zhao CJ (2019). Characterization and expression analysis of GPAT gene family in maize. Can J Plant Sci 99, 577-588. |
| [39] | Yue C, Cao HL, Wang L, Zhou YH, Huang YT, Hao XY, Wang YC, Wang B, Yang YJ, Wang XC (2015). Effects of cold acclimation on sugar metabolism and sugar-related gene expression in tea plant during the winter season. Plant Mol Biol 88, 591-608. |
| [40] | Zhang ZL, Liu ZH, Song HN, Chen MH, Cheng SP (2019). Protective role of leaf variegation in Pittosporum tobira under low temperature: insights into the physio-biochemical and molecular mechanisms. Int J Mol Sci 20, 4857. |
| [41] | Zhou Y, Zeng LT, Fu XM, Mei X, Cheng SH, Liao YY, Deng RF, Xu XL, Jiang YM, Duan XW, Susanne B, Yang ZY (2016). The sphingolipid biosynthetic enzyme Sphingolipid delta8 desaturase is important for chilling resistance of tomato. Sci Rep 6, 38742. |
| [1] | LU Ming-Hui LOU Qun-Feng CHEN Jin-Feng②. A Review on Chilling Injury and Cold Tolerance in Cucumis sativus L. [J]. Chinese Bulletin of Botany, 2004, 21(05): 578-586. |
| [2] | Yuan Yu-xin, Wang Ying, Pei Bao-hua, Jia Xu-guang. Effect of Cold Inducement on Hardiness of Amorpha fruticosa and Robinia pseudoacacia [J]. Chin J Plan Ecolo, 1996, 20(1): 65-73. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||