

Chinese Bulletin of Botany ›› 2022, Vol. 57 ›› Issue (3): 327-339.DOI: 10.11983/CBB21192 cstr: 32102.14.CBB21192
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Qiong Zhai, Rongqin Chen, Xiaohua Liang, Chuchun Zeng, Bo Hu, Ling Li, Xiaoyun Li( )
)
Received:2021-11-13
Accepted:2022-03-03
Online:2022-05-01
Published:2022-05-18
Contact:
Xiaoyun Li
About author:First author contact:† These authors contributed equally to this paper
Qiong Zhai, Rongqin Chen, Xiaohua Liang, Chuchun Zeng, Bo Hu, Ling Li, Xiaoyun Li. Establishment and Application of a Rapid Genetic Transformation Method for Peanut[J]. Chinese Bulletin of Botany, 2022, 57(3): 327-339.

Figure 2 The process of rapid genetic transformation in peanut (A) Cut off the top at the second stem node; (B) Preparation before transformation (bacteria, active agent and syringe, active agent is 100 μmol∙L-1 acetosyringone); (C) Syringe draws Agrobacterium; (D) Agrobacterium is injected at the second stem node of the peanut; (E) Dark culture for 3 to 5 days; (F) New buds grow; (G) Cut off the lateral bud; (H) Normal light culture; (I) Transplanting and replanting, the injection point should be covered with the soil; (J) Pick pods above the injection point (the white arrow indicated the injection point). Bars=1 cm
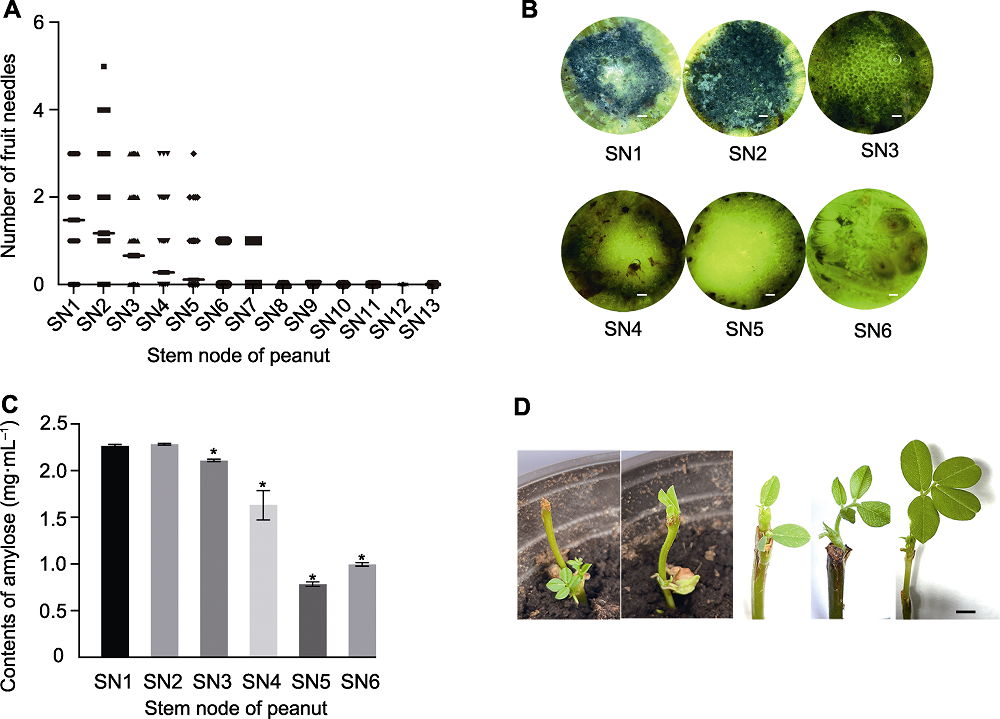
Figure 3 Selection of injection site for Agrobacterium in peanut (A) The 120 peanuts were randomly selected to calculate the occurrence site of gynophores in experimental farm; (B) Peanut stems were hand out sliced and stained with iodine solution, then take photos (bars=1 mm); (C) Measurement of starch content in each stem node (* represent the significant differences when compared with the first stem node (P<0.01)); (D) New shoots growth after Agrobacterium injection (bar=1 cm). SN1 represents the first stem node that occurred with two cotyledons, SN2 represents the second stem node that above the cotyledons, and so on.

Figure 4 Screening and identification of rapidly-transformation peanuts (A) The electrophoresis result is identification of pDR5:RUBY transgenic peanut plants through PCR using specific primers (R1-R5 represents independent transgenic lines; Mock, injected only with empty bacteria, the same as below); (B) Control (Mock) and pDR5:RUBY transgenic peanut leaves were observed by stereomicroscopy (bars=5 mm); (C) Phenotypic observation of T0 generation transgenic peanut, including control (Mock), pDR5:RUBY and p35S:GFP plants (bars=1 cm); (D) The electrophoresis result is the identification of p35S:GFP transgenic peanut plants using specific PCR primers (1-15 represent independent transgenic lines); (E) The fluorescence of GFP protein (green) were observed by stereomicroscopy in leaves of T0 p35S:GFP transgenic peanut, Mock is control plant, p35S:GFP is GFP overexpression plant (bars=50 μm); (F) Screening of T1 p35S:GFP transgenic peanuts using Kan resistance (bar=1 cm); (G) Identification of p35S:GFP transgenic peanut plants through Western Blot using GFP antibody (G1-G7 represents independent transgenic lines); (H) The fluorescence of GFP protein (green) were observed in T1 p35S:GFP protoplast by Confocal microscopy, red is chlorophyll a spontaneous fluorescence (bars=5 μm).
| Gene (species) | Generation | Total identified plants | Positive transgenic plants | Transformation rate (%) |
|---|---|---|---|---|
| p35S:GFP | T0 | 30 | 18 | 60 |
| pDR5:RUBY | T0 | 21 | 9 | 42.8 |
Table 1 The ratio of rapid transformation in peanut
| Gene (species) | Generation | Total identified plants | Positive transgenic plants | Transformation rate (%) |
|---|---|---|---|---|
| p35S:GFP | T0 | 30 | 18 | 60 |
| pDR5:RUBY | T0 | 21 | 9 | 42.8 |
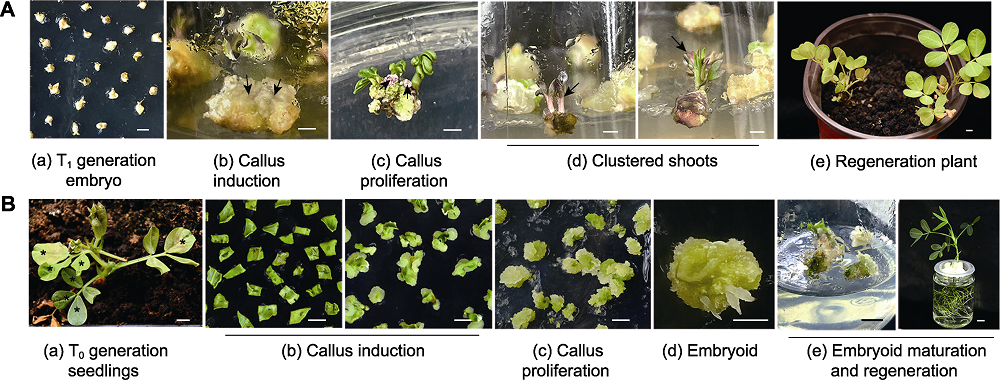
Figure 5 Purification and propagation of rapidly-transformation peanuts (A) Screening the embryo of T1 generation pDR5:RUBY transgenic peanut using hygromycin B resistance, then the callus and clustered shoots were induced from these resistance embryos, the transgenic plants were propagation using clustered shoots (the arrow indicated the high expression of pDR5:RUBY); (B) Transgenic plants were screening, purifying and propagating using tissue culture, these tissues came from peanut leaves that displayed the Basta resistance T0 plants (the * in (a) showed the Basta-resistance leaves). Bars=1 cm
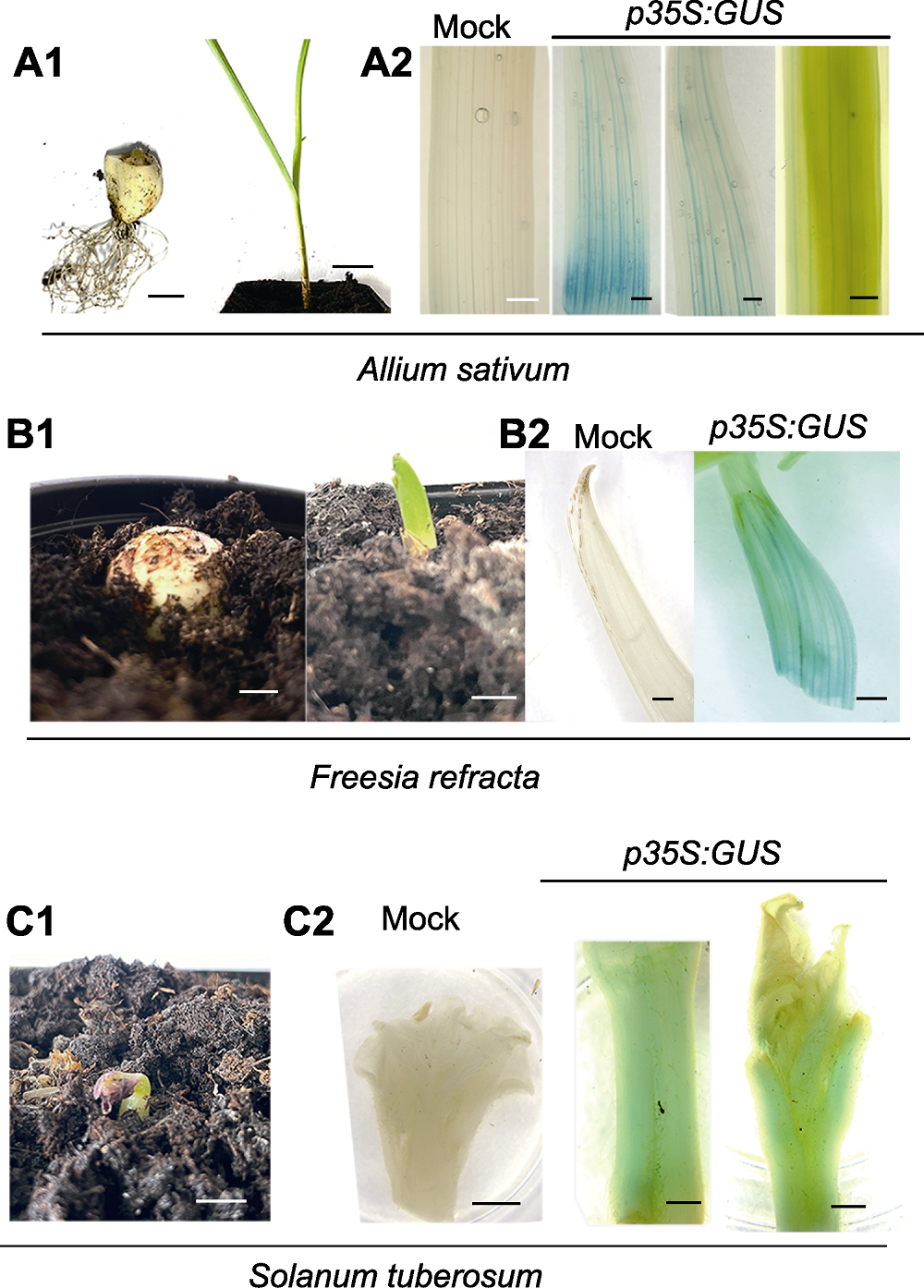
Figure 6 Rapidly-transformation of p35S:GUS in sprout sections from garlic, freesia and potato (A1) Garlic bud point section (left) and newly grown garlic after injection (right) (bars=1 cm); (A2) GUS detection of control (Mock) and p35S:GUS transgenic lines (bars=1 mm); (B1) Shoot spot section of freesia (left), and the new shoots after injection (right) (bars=1 cm); (B2) GUS detection of control (Mock) and p35S:GUS transgenic lines (bars=1 mm); (C1) The shoots of potato after injection (bar=1 cm); (C2) GUS detection of control (Mock, bar=1 cm) and p35S:GUS transgenic lines (bars=1 mm).
| [1] | 陈容钦, 李晓云, 李玲, 胡博 (2020). 一种花生原生质体提取方法及应用. 中国专利. CN111019879A. 2020-04-17. |
| [2] | 陈雪涛, 熊尚文, 袁雪梅, 张庆华 (2017). 上高紫皮大蒜品种退化原因及防止措施. 安徽农学通报 23(1), 51, 98. |
| [3] | 程秀云, 罗建玲, 罗玉芳 (2013). 生熟大蒜的抑菌作用研究. 食品与机械 29(6), 89-92. |
| [4] | 杜鹏飞, 王玉, 曹英萍, 杨松, 孙志超, 毛德才, 鄢家俊, 李达旭, 孙美贞, 付春祥, 白史且 (2021). 基因枪介导的老芒麦遗传转化体系的建立. 植物学报 56, 62-70. |
| [5] | 和小燕 (2017). 花生叶色突变体和AhPDS基因CRISPR- Cas9体系构建与遗传转化. 硕士论文. 郑州: 河南农业大学. pp. 42-43. |
| [6] | 李家磊, 管立军, 王崑仑, 高扬, 严松, 张志宏, 卢淑雯, 周野 (2020). 双波长法测定冰温贮藏西洋参中直链淀粉和支链淀粉的含量. 食品工业科技 41, 223-227. |
| [7] | 刘璨, 苏良辰, 庄依娜, 覃铭, 李玲 (2010). 花生幼叶不定芽诱导与快速繁殖. 亚热带植物科学 39, 21-24, 37. |
| [8] | 苗利娟, 黄冰艳, 张新友, 董文召, 刘娟, 秦利, 张俊, 齐飞艳, 石磊 (2017). 花生组培再生及农杆菌介导遗传转化研究进展. 中国农学通报 33(32), 15-20. |
| [9] | 彭振英, 单雷, 张智猛, 李新国, 万书波 (2019). 花生远缘杂交后代的氨基酸含量变异分析. 花生学报 48(1), 21-26. |
| [10] | 覃铭, 胡博, 刘璨, 李玲, 罗虹 (2010). AhNCED1基因转化花生研究. 热带亚热带植物学报 18, 277-282. |
| [11] | 徐静, 张新友, 汤丰收, 董文召, 臧秀旺, 张忠信, 韩锁义, 秦利 (2014). 花生新品种远杂9847选育及启示. 河南农业科学 43(10), 38-41. |
| [12] | 徐平丽, 唐桂英, 毕玉平, 柳展基, 单雷 (2018). 花生AhFAD2基因抑制表达的转基因后代分析. 生物工程学报 34(9), 1469-1477. |
| [13] |
徐悦, 曹英萍, 王玉, 付春祥, 戴绍军 (2019). 发根农杆菌介导的菠菜毛状根遗传转化体系的建立. 植物学报 54, 515- 521.
DOI |
| [14] | 杨澜, 刘雅, 项阳, 孙秀娟, 颜景畏, 张阿英 (2021). 谷子茎尖体外遗传转化体系的建立与优化. 植物学报 56, 71-79. |
| [15] | 殷冬梅 (2020). 高产高抗青枯病花生新品种农大花108. 中国种业 (11), 117-118. |
| [16] | 禹山林 (2008). 中国花生品种及其系谱. 上海: 上海科学技术出版社. pp. 233-745. |
| [17] | 张新友, 杜培, 刘华, 董文召, 秦利, 孙子淇, 徐静, 韩锁义, 张忠信, 苗利娟, 齐飞艳, 李丽娜, 付留洋, 王思雨, 房元瑾 (2019). 花生野生种质评价与利用技术创新及新品种培育. 河南省农业科学院经济作物研究所. 国家科技成果. 2019-12-01. |
| [18] |
Anuradha TS, Jami SK, Datla RS, Kirti PB (2006). Genetic transformation of peanut (Arachis hypogaea L.) using cotyledonary node as explant and a promoterless gus:: nptII fusion gene based vector. J Biosci 31, 235-246.
DOI URL |
| [19] |
Bakhshy E, Zarinkamar F, Nazari M (2020). Structural and quantitative changes of starch in seed of Trigonella persica during germination. Int J Biol Macromol 164, 1284- 1293.
DOI PMID |
| [20] |
Banavath JN, Chakradhar T, Pandit V, Konduru S, Guduru KK, Akila CS, Podha S, Puli COR (2018). Stress inducible overexpression of AtHDG11 leads to improved drought and salt stress tolerance in peanut (Arachis hypogaea L.). Front Chem 6, 34.
DOI PMID |
| [21] | Bhalani H, Thankappan R, Mishra GP, Sarkar T, Bosamia TC, Dobaria JR (2019). Regulation of antioxidant mechanisms by AtDREB1A improves soil-moisture deficit stress tolerance in transgenic peanut (Arachis hypogaea L.). PLoS One 14, e0216706. |
| [22] |
Bouchez D, Camilleri C (1990). Identification of a putative rol B gene on the TR-DNA of the Agrobacterium rhizogens A4 Ri plasmid. Plant Mol Biol 14, 617-619.
PMID |
| [23] |
Chen M, Yang Q, Wang T, Chen N, Pan L, Chi X, Yang ZH, Wang M, Yu S (2015). Agrobacterium-mediated genetic transformation of peanut and the efficient recovery of transgenic plants. Can J Plant Sci 95, 735-744.
DOI URL |
| [24] |
Ditt RF, Nester EW, Comai L (2001). Plant gene expression response to Agrobacterium tumefaciens. Proc Natl Acad Sci USA 98, 10954-10959.
DOI URL |
| [25] |
Gantait S, Mondal S (2018). Transgenic approaches for genetic improvement in groundnut (Arachis hypogaea L.) against major biotic and abiotic stress factors. J Genet Eng Biotechnol 16, 537-544.
DOI PMID |
| [26] |
Gayral M, Fanuel M, Rogniaux H, Dalgalarrondo M, Elmorjani K, Bakan B, Marion D (2019). The spatiotemporal deposition of lysophosphatidylcholine within starch granules of maize endosperm and its relationships to the expression of genes involved in endoplasmic reticulum- amyloplast lipid trafficking and galactolipid synthesis. Plant Cell Physiol 60, 139-151.
DOI URL |
| [27] |
He Y, Zhang T, Sun H, Zhan H, Zhao Y (2020). A reporter for noninvasively monitoring gene expression and plant transformation. Hortic Res 7, 152.
DOI URL |
| [28] |
Hu JC, Li SY, Li ZL, Li HY, Song WB, Zhao HM, Lai JS, Xia LQ, Li DW, Zhang YL (2019). A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol Plant Pathol 20, 1463-1474.
DOI URL |
| [29] |
Kiranmai K, Rao GL, Pandurangaiah M, Nareshkumar A, Reddy VA, Lokesh U, Venkatesh B, Johnson AMA, Sudhakar C (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front Plant Sci 9, 346.
DOI PMID |
| [30] |
Krishna G, Singh BK, Kim EK, Morya VK, Ramteke PW (2015). Progress in genetic engineering of peanut (Arachis hypogaea L.)—a review. Plant Biotechnol J 13, 147- 162.
DOI PMID |
| [31] |
Li LM, Zhang Z, Pan SY, Li L, Li XY (2019). Characterization and metabolism effect of seed endophytic bacteria associated with peanut grown in South China. Front Microbiol 10, 2659.
DOI URL |
| [32] |
Li TD, Hu JC, Sun Y, Li BS, Zhang DL, Li WL, Liu JX, Li DW, Gao CX, Zhang YL, Wang YP (2021). Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol Plant 14, 1787- 1798.
DOI URL |
| [33] |
Li YQ, Shan XT, Gao RF, Han TT, Zhang J, Wang YN, Kimani S, Wang L, Gao X (2020). MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Commun Biol 3, 396.
DOI URL |
| [34] |
Liu H, Hu DX, Du PX, Wang LP, Liang XQ, Li HF, Lu Q, Li SX, Liu HY, Chen XP, Varshney RK, Hong YB (2021a). Single-cell RNA-seq describes the transcriptome landscape and identifies critical transcription factors in the leaf blade of the allotetraploid peanut (Arachis hypogaea L.). Plant Biotechnol J 19, 2261-2276.
DOI URL |
| [35] |
Liu SJ, Liu CJ, Wang X, Chen HQ (2021b). Seed-specific activity of the Arabidopsis β-glucosidase 19 promoter in transgenic Arabidopsis and tobacco. Plant Cell Rep 40, 213-221.
DOI URL |
| [36] |
Liu X, Li LM, Li MJ, Su LC, Lian SM, Zhang BH, Li XY, Ge K, Li L (2018). AhGLK1 affects chlorophyll biosynthesis and photosynthesis in peanut leaves during recovery from drought. Sci Rep 8, 2250.
DOI URL |
| [37] |
Liu X, Li LM, Zhang BH, Zeng LD, Li L (2020). AhHDA1-mediated AhGLK1 promoted chlorophyll synthesis and photosynthesis regulates recovery growth of peanut leaves after water stress. Plant Sci 294, 110461.
DOI URL |
| [38] |
Lokesh U, Venkatesh B, Kiranmai K, Nareshkumar A, Amarnathareddy V, Rao GL, Johnson AMA, Pandurangaiah M, Sudhakar C (2019). Overexpression of ß-Ketoacyl Co-A Synthase1 gene improves tolerance of drought susceptible groundnut (Arachis hypogaea L.) cultivar K-6 by increased leaf epicuticular wax accumulation. Front Plant Sci 9, 1869.
DOI URL |
| [39] |
Manjulatha M, Sreevathsa R, Kumar AM, Sudhakar C, Prasad TG, Tuteja N, Udayakumar M (2014). Overexpression of a pea DNA helicase (PDH45) in peanut (Arachis hypogaea L.) confers improvement of cellular level tolerance and productivity under drought stress. Mol Biotechnol 56, 111-125.
DOI PMID |
| [40] |
Mauro ML, Bettini PP (2021). Agrobacterium rhizogenes rolB oncogene: an intriguing player for many roles. Plant Physiol Biochem 165, 10-18.
DOI URL |
| [41] |
Mehta R, Radhakrishnan T, Kumar A, Yadav R, Dobaria JR, Thirumalaisamy PP, Jain RK, Chigurupati P (2013). Coat protein-mediated transgenic resistance of peanut (Arachis hypogaea L.) to peanut stem necrosis disease through Agrobacterium-mediated genetic transformation. Indian J Virol 24, 205-213.
DOI URL |
| [42] |
Nemoto K, Hara M, Suzuki M, Seki H, Oka A, Muranaka T, Mano Y (2009). Function of the aux and rol genes of the Ri plasmid in plant cell division in vitro. Plant Signal Behav 4, 1145-1147.
DOI URL |
| [43] |
Pandurangaiah M, Rao GL, Sudhakarbabu O, Nareshkumar A, Kiranmai K, Lokesh U, Thapa G, Sudhakar C (2014). Overexpression of horsegram (Macrotyloma uniflorum Lam.Verdc.) NAC transcriptional factor (MuNAC4) in groundnut confers enhanced drought tolerance. Mol Biotechnol 56, 758-769.
DOI PMID |
| [44] |
Penfield S (2017). Seed dormancy and germination. Curr Biol 27, 874-878.
DOI PMID |
| [45] |
Su LC, Liu S, Liu X, Zhang BH, Li MJ, Zeng LD, Li L (2021). Transcriptome profiling reveals histone deacetylase 1 gene overexpression improves flavonoid, isoflavonoid, and phenylpropanoid metabolism in Arachis hypogaea hairy roots. Peer J 9, e10976.
DOI URL |
| [46] |
Tang GY, Xu PL, Ma WH, Wang F, Liu ZJ, Wan SB, Shan L (2018). Seed-specific expression of AtLEC1increased oil content and altered fatty acid composition in seeds of peanut (Arachis hypogaea L.). Front Plant Sci 9, 260
DOI URL |
| [47] |
Tiwari S, Mishra DK, Singh A, Singh PK, Tuli R (2008). Expression of a synthetic cry1EC gene for resistance against Spodoptera litura in transgenic peanut (Arachis hypogaea L.). Plant Cell Rep 27, 1017-1025.
DOI URL |
| [48] | Tiwari V, Chaturvedi AK, Mishra A, Jha B (2015). Introgression of the SbASR-1 gene cloned from a halophyte Salicornia brachiata enhances salinity and drought endurance in transgenic groundnut (Arachis hypogaea) and acts as a transcription factor. PLoS One 10, e0131567. |
| [49] |
Yan Y, Zhu XJ, Yu Y, Li C, Zhang ZL, Wang F (2022). Nanotechnology strategies for plant genetic engineering. Adv Mater 34, 2106945.
DOI URL |
| [50] |
Zhang CZ, Yang ZM, Tang D, Zhu YH, Wang P, Li DW, Zhu GT, Xiong XY, Shang Y, Li CH, Huang SW (2021). Genome design of hybrid potato. Cell 184, 3873-3883.
DOI URL |
| [51] |
Zhao M, Zhang HX, Yan H, Qiu L, Baskin CC (2018). Mobilization and role of starch, protein, and fat reserves during seed germination of six wild grassland species. Front Plant Sci 9, 234.
DOI URL |
| [52] | Zhuang WJ, Chen H, Yang M, Wang JP, Pandey MK, Zhang C, Chang WC, Zhang LS, Zhang XT, Tang RH, Garg V, Wang XJ, Tang HB, Chow CN, Wang JP, Deng Y, Wang DP, Khan AW, Yang Q, Cai TC, Bajaj P, Wu KC, Guo BZ, Zhang XY, Li JJ, Liang F, Hu J, Liao BS, Liu SY, Chitikineni A, Yan HS, Zheng YX, Shan SH, Liu QZ, Xie DY, Wang ZY, Khan SA, Ali N, Zhao CZ, Li XG, Luo ZL, Zhang SB, Zhuang RR, Peng Z, Wang SY, Mamadou G, Zhuang YH, Zhao ZF, Yu WC, Xiong FQ, Quan WP, Yuan M, Li Y, Zou HS, Xia H, Zha L, Fan JP, Yu JG, Xie WP, Yuan JQ, Chen K, Zhao SS, Chu WT, Chen YT, Sun PC, Meng FB, Zhuo T, Zhao YH, Li CJ, He GH, Zhao YL, Wang CC, Kavikishor PB, Pan RL, Paterson AH, Wang XY, Ming R, Varshney RK (2019). The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat Genet 51, 865-876. |
| [1] | Mei Yan, Xinyou Zhang, Suoyi Han, Bingyan Huang, Wenzhao Dong, Hua Liu, Ziqi Sun. Genome-wide Association Study of Agronomic and Yield Traits in a Worldwide Collection of Peanut (Arachis hypogaea) Germplasm [J]. Chinese Bulletin of Botany, 2015, 50(4): 460-472. |
| [2] | ZHANG Jia-Lei,LI Xiang-Dong,YANG Chuan-Ting,GAO Fang,ZHANG Feng,WANG Yuan-Yuan,SUN Lian-Qiang. Relationship of quality formation and ultrastructure of cotyledon cells in two quality types of peanut [J]. Chin J Plant Ecol, 2013, 37(8): 768-776. |
| [3] | CI Dun-Wei,DAI Liang-Xiang,SONG Wen-Wu,ZHANG Zhi-Meng. Genotypic differences in salt tolerance from germination to seedling stage in peanut [J]. Chin J Plant Ecol, 2013, 37(11): 1018-1027. |
| [4] | ZHANG Zhi-Meng, WAN Shu-Bo, DAI Liang-Xiang, SONG Wen-Wu, CHEN Jing, SHI Yun-Qing. Estimating and screening of drought resistance indexes of peanut [J]. Chin J Plant Ecol, 2011, 35(1): 100-109. |
| [5] | LI Hua-Shou, ZHANG Xiu-Yu, ZENG Xiang-You, NIE Cheng-Rong. TOXIC EFFECTS OF POTASSIUM CHLORATE ON PEANUT GROWTH [J]. Chin J Plant Ecol, 2006, 30(1): 124-131. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||